Pairwise distances: Difference between revisions
Jump to navigation
Jump to search
mNo edit summary |
(https://github.com/schrodinger/pymol-open-source/issues/20) |
||
| (One intermediate revision by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = script | |type = script | ||
|download = | |download = [http://dx.doi.org/10.6084/m9.figshare.1031600 figshare] | ||
|author = [[User:PietroGattiLafranconi|Pietro Gatti-Lafranconi]] | |author = [[User:PietroGattiLafranconi|Pietro Gatti-Lafranconi]] | ||
|license = [http://creativecommons.org/licenses/by | |license = [http://creativecommons.org/licenses/by/4.0/ CC BY 4.0] | ||
}} | }} | ||
| Line 87: | Line 87: | ||
<source lang="python"> | <source lang="python"> | ||
from __future__ import print_function | |||
from pymol import cmd, stored, math | from pymol import cmd, stored, math | ||
| Line 99: | Line 100: | ||
show: shows (Y) individual distances in pymol menu (default=N) | show: shows (Y) individual distances in pymol menu (default=N) | ||
""" | """ | ||
print "" | print("") | ||
cmd.delete ("dist*") | cmd.delete("dist*") | ||
extra="" | extra="" | ||
if sidechain=="Y": extra=" and not name c+o+n" | if sidechain=="Y": | ||
extra=" and not name c+o+n" | |||
#builds models | #builds models | ||
m1=cmd.get_model(sel2+" around "+str(max_dist)+" and "+sel1+extra) | m1 = cmd.get_model(sel2+" around "+str(max_dist)+" and "+sel1+extra) | ||
m1o=cmd.get_object_list(sel1) | m1o = cmd.get_object_list(sel1) | ||
m2=cmd.get_model(sel1+" around "+str(max_dist)+" and "+sel2+extra) | m2 = cmd.get_model(sel1+" around "+str(max_dist)+" and "+sel2+extra) | ||
m2o=cmd.get_object_list(sel2) | m2o = cmd.get_object_list(sel2) | ||
#defines selections | #defines selections | ||
| Line 120: | Line 122: | ||
#controlers-1 | #controlers-1 | ||
if len(m1o)==0: | if len(m1o)==0: | ||
print "warning, '"+sel1+extra+"' does not contain any atoms." | print("warning, '"+sel1+extra+"' does not contain any atoms.") | ||
return | return | ||
if len(m2o)==0: | if len(m2o)==0: | ||
print "warning, '"+sel2+extra+"' does not contain any atoms." | print("warning, '"+sel2+extra+"' does not contain any atoms.") | ||
return | return | ||
| Line 139: | Line 141: | ||
#controler-2 | #controler-2 | ||
if counter==0: | if counter==0: | ||
print "warning, no distances were measured! Check your selections/max_dist value" | print("warning, no distances were measured! Check your selections/max_dist value") | ||
return | return | ||
#outputs | #outputs | ||
if output=="S": print s | if output=="S": | ||
print(s) | |||
if output=="P": | if output=="P": | ||
f=open('IntAtoms_'+max_dist+'.txt','w') | f=open('IntAtoms_'+max_dist+'.txt','w') | ||
| Line 149: | Line 152: | ||
f.write(s) | f.write(s) | ||
f.close() | f.close() | ||
print "Results saved in IntAtoms_%s.txt" % max_dist | print("Results saved in IntAtoms_%s.txt" % max_dist) | ||
print "Number of distances calculated: %s" % (counter) | print("Number of distances calculated: %s" % (counter)) | ||
cmd.hide("lines", "IntRes_*") | cmd.hide("lines", "IntRes_*") | ||
if show=="Y": cmd.show("lines","IntRes_"+max_dist) | if show=="Y": cmd.show("lines","IntRes_"+max_dist) | ||
Latest revision as of 15:03, 12 March 2019
| Type | Python Script |
|---|---|
| Download | figshare |
| Author(s) | Pietro Gatti-Lafranconi |
| License | CC BY 4.0 |
Introduction
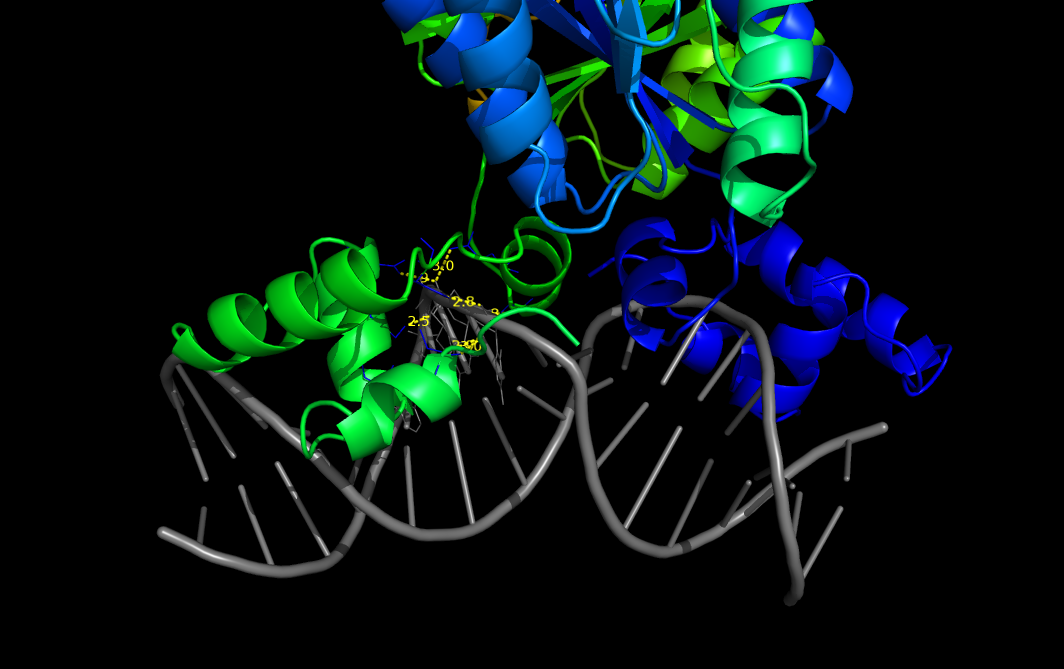
Given any two selections, this script calculates and returns the pairwise distances between all atoms that fall within a defined distance.
Can be used to measure distances within the same chain, between different chains or different objects.
Distances can be restricted to sidechain atoms only and the outputs either displayed on screen or printed on file.
Usage
pairwise_dist sel1, sel2, max_dist, [output=S/P/N, [sidechain=N/Y, [show=Y/N]]]
Required Arguments
- sel1 = first selection
- sel2 = second selection
- max_dist = max distance in Angstroms
Optional Arguments
- output = accepts Screen/Print/None (default N)
- sidechain = limits (Y) results to sidechain atoms (default N)
- show = shows (Y) individual distances in pymol menu (default=N)
Examples
example #1
PyMOL>pairwise_dist 1efa and chain D, 1efa and chain B, 3, output=S, show=Y
1efa/D/DC/13/OP1 to 1efa/B/TYR/47/OH: 2.765
1efa/D/DC/13/OP2 to 1efa/B/LEU/6/N: 2.983
1efa/D/DC/13/OP2 to 1efa/B/LEU/6/CB: 2.928
1efa/D/DT/14/O4' to 1efa/B/ALA/57/CB: 2.827
1efa/D/DT/14/OP1 to 1efa/B/ASN/25/OD1: 2.858
1efa/D/DT/14/OP1 to 1efa/B/GLN/54/NE2: 2.996
1efa/D/DT/14/OP2 to 1efa/B/SER/21/OG: 2.517
1efa/D/DC/15/N4 to 1efa/B/GLN/18/NE2: 2.723
1efa/D/DA/16/N6 to 1efa/B/GLN/18/NE2: 2.931
Number of distances calculated: 9
example #2
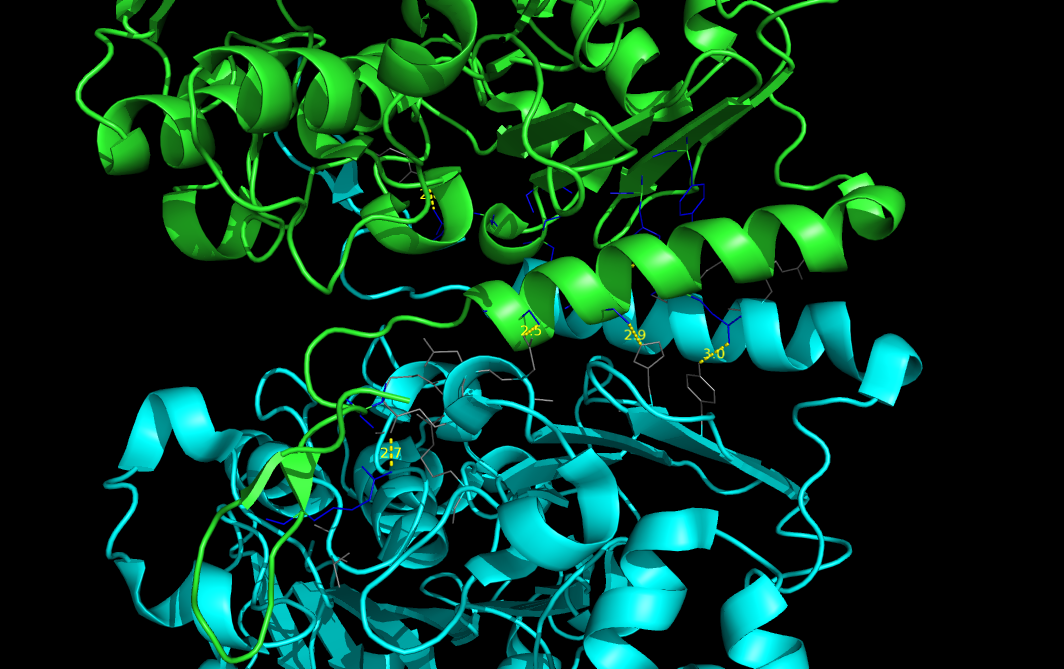
PyMOL>pairwise_dist 2w8s and chain a, 2W8s and chain b, 3, sidechain=Y, output=S, show=Y
2W8S/A/GLN/432/OE1 to 2W8S/B/ARG/503/NH2: 2.758
2W8S/A/ASP/434/OD1 to 2W8S/B/SER/493/OG: 2.444
2W8S/A/TYR/447/OH to 2W8S/B/GLN/485/NE2: 2.878
2W8S/A/HIS/449/NE2 to 2W8S/B/SER/489/OG: 2.686
2W8S/A/GLN/485/OE1 to 2W8S/B/TYR/447/OH: 2.971
2W8S/A/SER/489/OG to 2W8S/B/HIS/449/NE2: 2.913
2W8S/A/SER/493/OG to 2W8S/B/ASP/434/OD1: 2.491
2W8S/A/ARG/503/NH2 to 2W8S/B/GLN/432/OE1: 2.653
Number of distances calculated: 8
example #3
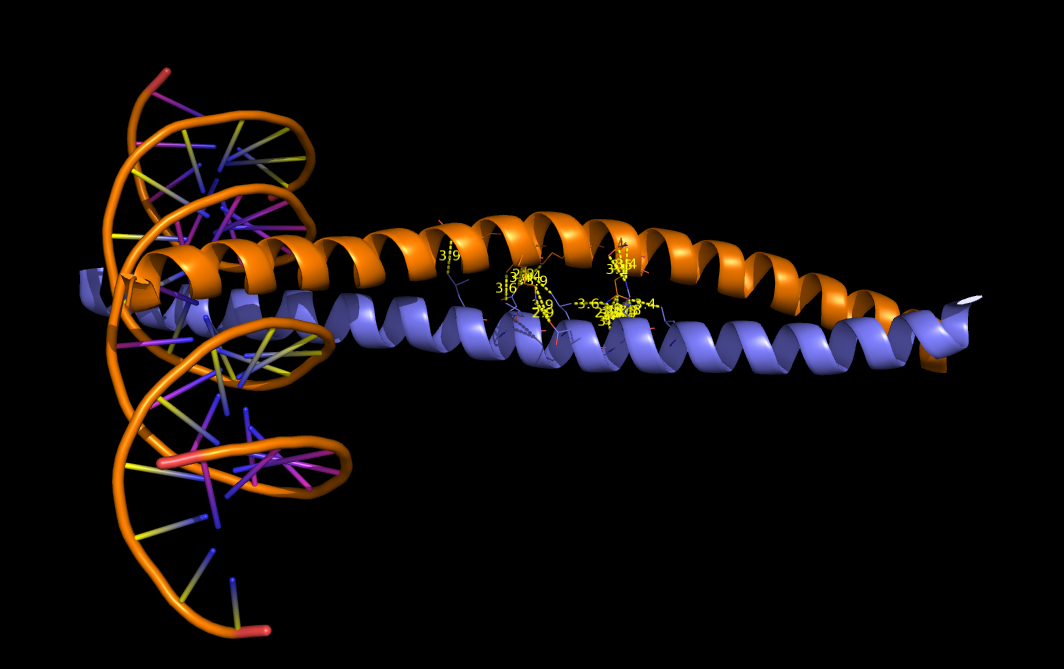
PyMOL>pairwise_dist 1FOS and chain H and resi 290:300, 1FOS and chain G, 4, sidechain=Y, show=Y, output=P
Results saved in IntAtoms_4.txt
Number of distances calculated: 24
The Code
Copy the following text and save it as pairwisedistances.py
from __future__ import print_function
from pymol import cmd, stored, math
def pairwise_dist(sel1, sel2, max_dist, output="N", sidechain="N", show="N"):
"""
usage: pairwise_dist sel1, sel2, max_dist, [output=S/P/N, [sidechain=N/Y, [show=Y/N]]]
sel1 and sel2 can be any to pre-existing or newly defined selections
max_dist: maximum distance in Angstrom between atoms in the two selections
--optional settings:
output: accepts Screen/Print/None (default N)
sidechain: limits (Y) results to sidechain atoms (default N)
show: shows (Y) individual distances in pymol menu (default=N)
"""
print("")
cmd.delete("dist*")
extra=""
if sidechain=="Y":
extra=" and not name c+o+n"
#builds models
m1 = cmd.get_model(sel2+" around "+str(max_dist)+" and "+sel1+extra)
m1o = cmd.get_object_list(sel1)
m2 = cmd.get_model(sel1+" around "+str(max_dist)+" and "+sel2+extra)
m2o = cmd.get_object_list(sel2)
#defines selections
cmd.select("__tsel1a", sel1+" around "+str(max_dist)+" and "+sel2+extra)
cmd.select("__tsel1", "__tsel1a and "+sel2+extra)
cmd.select("__tsel2a", sel2+" around "+str(max_dist)+" and "+sel1+extra)
cmd.select("__tsel2", "__tsel2a and "+sel1+extra)
cmd.select("IntAtoms_"+max_dist, "__tsel1 or __tsel2")
cmd.select("IntRes_"+max_dist, "byres IntAtoms_"+max_dist)
#controlers-1
if len(m1o)==0:
print("warning, '"+sel1+extra+"' does not contain any atoms.")
return
if len(m2o)==0:
print("warning, '"+sel2+extra+"' does not contain any atoms.")
return
#measures distances
s=""
counter=0
for c1 in range(len(m1.atom)):
for c2 in range(len(m2.atom)):
distance=math.sqrt(sum(map(lambda f: (f[0]-f[1])**2, zip(m1.atom[c1].coord,m2.atom[c2].coord))))
if distance<float(max_dist):
s+="%s/%s/%s/%s/%s to %s/%s/%s/%s/%s: %.3f\n" % (m1o[0],m1.atom[c1].chain,m1.atom[c1].resn,m1.atom[c1].resi,m1.atom[c1].name,m2o[0],m2.atom[c2].chain,m2.atom[c2].resn,m2.atom[c2].resi,m2.atom[c2].name, distance)
counter+=1
if show=="Y": cmd.distance (m1o[0]+" and "+m1.atom[c1].chain+"/"+m1.atom[c1].resi+"/"+m1.atom[c1].name, m2o[0]+" and "+m2.atom[c2].chain+"/"+m2.atom[c2].resi+"/"+m2.atom[c2].name)
#controler-2
if counter==0:
print("warning, no distances were measured! Check your selections/max_dist value")
return
#outputs
if output=="S":
print(s)
if output=="P":
f=open('IntAtoms_'+max_dist+'.txt','w')
f.write("Number of distances calculated: %s\n" % (counter))
f.write(s)
f.close()
print("Results saved in IntAtoms_%s.txt" % max_dist)
print("Number of distances calculated: %s" % (counter))
cmd.hide("lines", "IntRes_*")
if show=="Y": cmd.show("lines","IntRes_"+max_dist)
cmd.deselect()
cmd.extend("pairwise_dist", pairwise_dist)