CMPyMOL: Difference between revisions
m (fix download url) |
|||
| (5 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = Software | |type = Software | ||
|download = | |download = https://github.com/emptyewer/CMPyMOL | ||
|version = 1.0b | |version = 1.0b | ||
|author = [[User:Venky|Venkatramanan Krishnamani]] | |author = [[User:Venky|Venkatramanan Krishnamani]] | ||
| Line 7: | Line 7: | ||
}} | }} | ||
CMPyMOL is an add-on software to molecular visualization program PyMOL. It combines the 3D visualization capabilities of PyMOL and 2D | CMPyMOL is an add-on software to molecular visualization program PyMOL. It combines the protein 3D visualization capabilities of PyMOL and the protein's 2D contact map with an interactive interface for scientific analysis. This software is freely distributed under the MIT license for Linux and Mac OS X platforms. | ||
== Website == | == Website == | ||
http://emptyewer.github.io/CMPyMOL/ | http://emptyewer.github.io/CMPyMOL/ | ||
== Version History == | |||
* Added support for reading multi-model PDB files. (Multi-Model PDB file for NMR structure or Trajectory from MD simulations.) | |||
* Supports displaying variance of contact points from a series of contact maps generated from multi-model PDB files. | |||
* CMPyMOL stores the calculated contact maps, heat maps and contact density information in a local SQLite database for fast and easy subsequent access. | |||
* Cleaner GUI. | |||
* Parallelized the code for contact map calculation for multi-frame PDB files. | |||
== Prerequisites == | == Prerequisites == | ||
| Line 21: | Line 29: | ||
** [http://www.pythonware.com/products/pil/ python imaging library (PIL)] | ** [http://www.pythonware.com/products/pil/ python imaging library (PIL)] | ||
** [http://www.numpy.org numpy] | ** [http://www.numpy.org numpy] | ||
* PyMOL | * PyMOL. (It is recommended that the user add the PyMOL installation directory to the $PATH environment variable.) | ||
* Stride | * Stride secondary structure assignment tool. This program can be downloaded from http://webclu.bio.wzw.tum.de/stride/ and compiled into a stand-alone executable. It is recommended that the Stride executable or its installation directory is added to the $PATH environment variable. NOTE: If this executable is not detected in the $PATH variable, the secondary structure calculation will be disabled in CMPyMOL. | ||
=== Mac OS X === | === Mac OS X === | ||
* Users can install the python libraries using "easy_install" or "pip". It is | * Users can install the python libraries using "easy_install" or "pip". It is recommended that the user use Enthought Canopy python distribution and management package downloaded from https://www.enthought.com/products/canopy/. This package includes a robust python library management software and a python IDE. | ||
* PyMOL 1.5.x can be installed using MacPorts http://www.macports.org. NOTE: This automatically adds the executable into $PATH. | * PyMOL 1.5.x can be installed using MacPorts http://www.macports.org. NOTE: This automatically adds the executable into the $PATH environment variable. | ||
=== Linux === | === Linux === | ||
* The python dependencies and PyMOL can be installed using apt-get (aptitude) or similar package management system. | * The python dependencies and PyMOL can be installed using apt-get (aptitude) or a similar package management system. | ||
== Installation == | == Installation == | ||
There is no need for installation of the script. Optionally, a standalone executable can be complied using "pyinstaller" or "py2exe" or "py2app" | There is no need for installation of the script. Optionally, a standalone executable can be complied using "pyinstaller" or "py2exe" or "py2app" package, depending on the users operating system. | ||
== Usage == | == Usage == | ||
<source lang="bash"> | <source lang="bash"> | ||
python CMPyMOL.py | python /<path to CMPyMOL directory>/CMPyMOL.py | ||
</source> | </source> | ||
| Line 46: | Line 54: | ||
== Software == | == Software == | ||
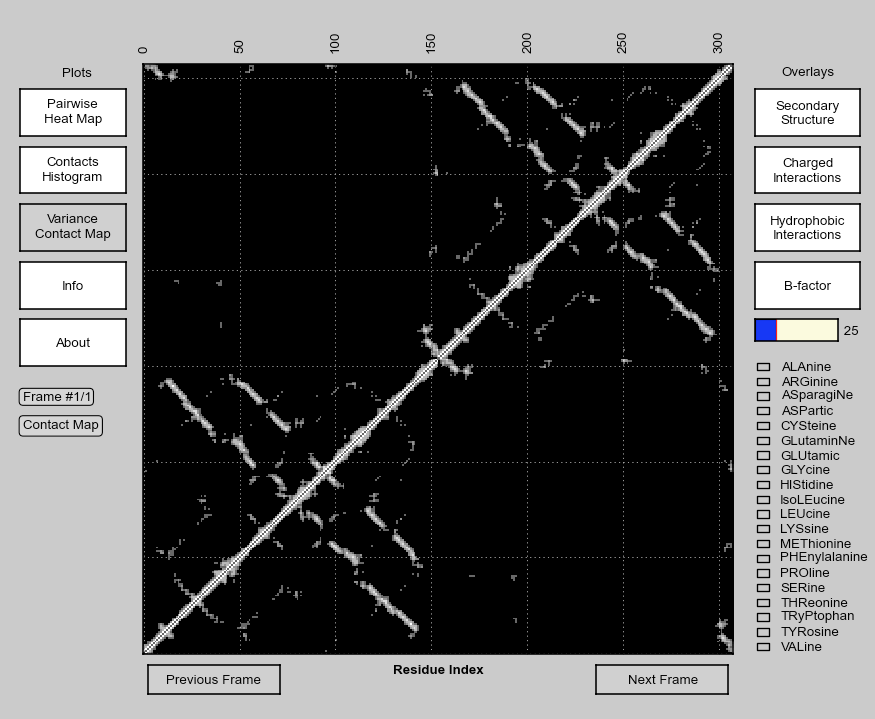
Clicking (left) and | Clicking (left) and dragging a selection of contact points on the displayed contact map will highlight the corresponding residues in the PyMOL window (as red and blue colored atoms in spheres representation). In addition, several structural/biochemical properties can be overlaid on top of the contact map. The contact-map data can also be plotted in other representations. The calculated contact-map, heat-map and contact density information is stored in a local SQL database. Any subsequent access of the same PDB with matching parameters will be read from the database for fast access. The code for calculating contact map for trajectory files is parallelized for efficiency. | ||
=== Inputs === | === Inputs === | ||
* Single-frame PDB files (local) | * Single-frame PDB files (local) | ||
* Multi-frame PDB trajectory files (local) | * Multi-frame PDB trajectory files (local) | ||
* Multi-frame trajectory files should have the following format | * Multi-frame trajectory files should have the following format. | ||
<source lang="bash"> | <source lang="bash"> | ||
| Line 65: | Line 73: | ||
ENDMDL | ENDMDL | ||
</source> | </source> | ||
NOTE: The PDB can include REMARKS, CRYST and other standard PDB information entries. The MODEL line is essential for the software to work properly (ENDMDL is optional). | |||
=== Overlays === | === Overlays === | ||
* Secondary structure of the protein as translucent strips over the contact map. This button won't be active if secondary structure calculation program stride is not | * Secondary structure of the protein is overlaid as translucent strips over the contact map. This button won't be active if secondary structure calculation program stride is not found in the system path ($PATH). (Button: Secondary Structure) | ||
* Contact points where a Charge-Charge interaction occurs. (Button: Charged Interactions) | * Contact points where a Charge-Charge interaction occurs are highlighted. (Button: Charged Interactions) | ||
* Residues that interact via hydrophobic interaction. (Button: Hydrophobic Interactions) | * Residues that interact via hydrophobic interaction are highlighted. (Button: Hydrophobic Interactions) | ||
* Contact regions that have a | * Contact regions that have a B-factor that is higher than a certain cutoff are highlighted (Button: B-factor). The b-factor cutoff can be varied using a slider (Slider). | ||
* Highlights a contact point/region where the | * Highlights a contact point/region where the pair of selected residues are in contact (selected by checking the checkboxes). If only one aminoacid is selected from the list, interaction site of the selected aminoacid with another one of the same type is highlighted. (List of checkboxes for each aminoacid) | ||
=== Plots === | === Plots === | ||
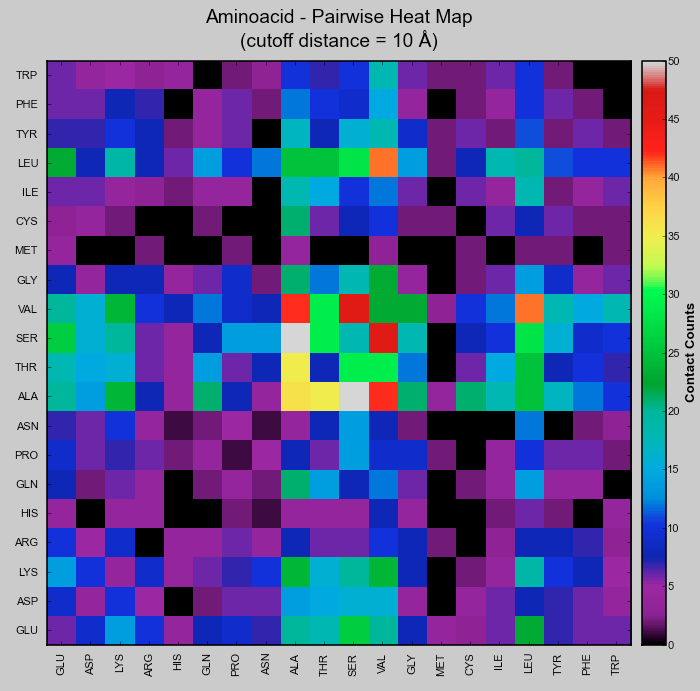
* Pairwise Heat Map - Plots a 20x20 matrix | * Pairwise Heat Map - Plots a 20x20 matrix of pairwise aminoacid interaction count. | ||
* Contacts Histogram - Plots the number of contacts | * Contacts Histogram - Plots the number of contacts around a given residue. Selecting a particular bar highlights the corresponding residue in the PyMOL window. | ||
* Variance Contact Map - For Multi-frame PDB files (trajectory), this button toggles the displays the variance | * Variance Contact Map - For Multi-frame PDB files (trajectory), this button toggles the displays the variance contact map starting from the initial frame until the current frame. This view can be used to identifying the dynamic regions in a protein. | ||
=== Word of Caution === | |||
When using a multi-frame PDB file, the contact-map for the next frame(s) are being pre-calculated in the background (depending on the number of free CPU cores available). Clicking on "Next Frame" in rapid succession may lead to undesired results and/or a crash. | |||
In the event of a crash, delete the database that is created in the working directory and relaunch the program. | |||
== Requests and Disclaimer == | == Requests and Disclaimer == | ||
Users are welcome to send me an email to request the addition of a specific feature or report a bug | Users are welcome to send me an email to request the addition of a specific feature or to report a bug. | ||
== | == Main Window == | ||
[[File:CMPyMOL.PNG|thumb|877px|center]] | [[File:CMPyMOL.PNG|thumb|877px|center]] | ||
== Pairwise Aminoacid Heatmap == | |||
[[File:Heatmap-CMPyMOL.PNG|thumb|877px|center]] | |||
== License == | == License == | ||
Latest revision as of 10:06, 5 May 2016
| Type | Software |
|---|---|
| Download | https://github.com/emptyewer/CMPyMOL |
| Author(s) | Venkatramanan Krishnamani |
| License | The MIT License (MIT) |
CMPyMOL is an add-on software to molecular visualization program PyMOL. It combines the protein 3D visualization capabilities of PyMOL and the protein's 2D contact map with an interactive interface for scientific analysis. This software is freely distributed under the MIT license for Linux and Mac OS X platforms.
Website
http://emptyewer.github.io/CMPyMOL/
Version History
- Added support for reading multi-model PDB files. (Multi-Model PDB file for NMR structure or Trajectory from MD simulations.)
- Supports displaying variance of contact points from a series of contact maps generated from multi-model PDB files.
- CMPyMOL stores the calculated contact maps, heat maps and contact density information in a local SQLite database for fast and easy subsequent access.
- Cleaner GUI.
- Parallelized the code for contact map calculation for multi-frame PDB files.
Prerequisites
- Python 2.7
- Python module dependency
- PyMOL. (It is recommended that the user add the PyMOL installation directory to the $PATH environment variable.)
- Stride secondary structure assignment tool. This program can be downloaded from http://webclu.bio.wzw.tum.de/stride/ and compiled into a stand-alone executable. It is recommended that the Stride executable or its installation directory is added to the $PATH environment variable. NOTE: If this executable is not detected in the $PATH variable, the secondary structure calculation will be disabled in CMPyMOL.
Mac OS X
- Users can install the python libraries using "easy_install" or "pip". It is recommended that the user use Enthought Canopy python distribution and management package downloaded from https://www.enthought.com/products/canopy/. This package includes a robust python library management software and a python IDE.
- PyMOL 1.5.x can be installed using MacPorts http://www.macports.org. NOTE: This automatically adds the executable into the $PATH environment variable.
Linux
- The python dependencies and PyMOL can be installed using apt-get (aptitude) or a similar package management system.
Installation
There is no need for installation of the script. Optionally, a standalone executable can be complied using "pyinstaller" or "py2exe" or "py2app" package, depending on the users operating system.
Usage
python /<path to CMPyMOL directory>/CMPyMOL.py
- This command will automatically invoke the PyMOL executable and the user is led through the rest of the program with a series of pop-up windows.
Software
Clicking (left) and dragging a selection of contact points on the displayed contact map will highlight the corresponding residues in the PyMOL window (as red and blue colored atoms in spheres representation). In addition, several structural/biochemical properties can be overlaid on top of the contact map. The contact-map data can also be plotted in other representations. The calculated contact-map, heat-map and contact density information is stored in a local SQL database. Any subsequent access of the same PDB with matching parameters will be read from the database for fast access. The code for calculating contact map for trajectory files is parallelized for efficiency.
Inputs
- Single-frame PDB files (local)
- Multi-frame PDB trajectory files (local)
- Multi-frame trajectory files should have the following format.
MODEL X
.
.
.
ATOM ...
ATOM ...
.
.
.
ENDMDL
NOTE: The PDB can include REMARKS, CRYST and other standard PDB information entries. The MODEL line is essential for the software to work properly (ENDMDL is optional).
Overlays
- Secondary structure of the protein is overlaid as translucent strips over the contact map. This button won't be active if secondary structure calculation program stride is not found in the system path ($PATH). (Button: Secondary Structure)
- Contact points where a Charge-Charge interaction occurs are highlighted. (Button: Charged Interactions)
- Residues that interact via hydrophobic interaction are highlighted. (Button: Hydrophobic Interactions)
- Contact regions that have a B-factor that is higher than a certain cutoff are highlighted (Button: B-factor). The b-factor cutoff can be varied using a slider (Slider).
- Highlights a contact point/region where the pair of selected residues are in contact (selected by checking the checkboxes). If only one aminoacid is selected from the list, interaction site of the selected aminoacid with another one of the same type is highlighted. (List of checkboxes for each aminoacid)
Plots
- Pairwise Heat Map - Plots a 20x20 matrix of pairwise aminoacid interaction count.
- Contacts Histogram - Plots the number of contacts around a given residue. Selecting a particular bar highlights the corresponding residue in the PyMOL window.
- Variance Contact Map - For Multi-frame PDB files (trajectory), this button toggles the displays the variance contact map starting from the initial frame until the current frame. This view can be used to identifying the dynamic regions in a protein.
Word of Caution
When using a multi-frame PDB file, the contact-map for the next frame(s) are being pre-calculated in the background (depending on the number of free CPU cores available). Clicking on "Next Frame" in rapid succession may lead to undesired results and/or a crash.
In the event of a crash, delete the database that is created in the working directory and relaunch the program.
Requests and Disclaimer
Users are welcome to send me an email to request the addition of a specific feature or to report a bug.
Main Window
Pairwise Aminoacid Heatmap
License
# The MIT License (MIT)
# =======================
#
# The PyMOL Plugin source code in this file is copyrighted, but you are
# free to use and copy it as long as you don't change or remove any of
# the copyright notices.
#
# -----------------------------------------------------------------------------------
# CMPyMOL
# Copyright (C) 2013 by Venkatramanan Krishnamani <venks@andrew.cmu.edu>
#
# Permission is hereby granted, free of charge, to any person obtaining a copy of this
# software and associated documentation files (the "Software"), to deal in the Software
# without restriction, including without limitation the rights to use, copy, modify,
# merge, publish, distribute, sublicense, and/or sell copies of the Software, and to
# permit persons to whom the Software is furnished to do so, subject to the following
# conditions:
#
# The above copyright notice and this permission notice shall be included in all copies
# or substantial portions of the Software.
#
# THE SOFTWARE IS PROVIDED "AS IS", WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED,
# INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR
# PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS BE
# LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT,
# TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE SOFTWARE OR THE USE OR
# OTHER DEALINGS IN THE SOFTWARE.