Cheshift: Difference between revisions
| (19 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = plugin | |type = plugin | ||
| | |download = https://github.com/aloctavodia/cheshift/archive/v3.6.zip | ||
|author = [[User:OsvaldoMartin|Osvaldo Martin]] | |author = [[User:OsvaldoMartin|Osvaldo Martin]] | ||
|license = GPL | |license = GPL | ||
| Line 8: | Line 8: | ||
== Description == | == Description == | ||
[[Image:cheshift.png|thumb|Result of a CheShift analysis | [[Image:cheshift.png|thumb|Result of a CheShift analysis.<br> | ||
Colors indicate the difference between predicted and observed <sup>13</sup>C<sup>α</sup> and/or <sup>13</sup>C<sup>β</sup>chemical shifts values averaged over all uploaded conformers. | |||
Green, yellow and red colors represent small, medium and large differences, respectively. White is used if either the prediction failed or the observed value is missing.<br> | |||
Blue is used to highlight residues for which the agreement between observed and predicted <sup>13</sup>C<sup>α</sup> and <sup>13</sup>C<sup>β</sup>chemical shifts can be improved, i.e., if the (χ<sup>1</sup> /χ<sup>2</sup> ) side-chain | 440px]] | |||
This plugin provides a way to use PyMOL to validate a protein model using observed chemical shifts | CheShift (pronounced /tʃeʃɪft/) is a software for prediction of 13Cα and 13Cβ chemical shifts and validation of proteins structures. It is based on the idea that the differences between observed and predicted <sup>13</sup>C<sup>α</sup> and <sup>13</sup>C<sup>β</sup> chemical shifts can be used as a sensitive probe with which to detect possible local flaws in protein structures. A [http://www.cheshift.com Web Server] is also available. | ||
This plugin provides a way to use PyMOL to validate a protein model using observed chemical shifts. | |||
=== Version === | === Version === | ||
The current version of this plugin is | The current version of this plugin is 3.6 | ||
== Installation == | == Installation == | ||
=== Linux === | === Linux === | ||
1) The plugin can be downloaded from here [[https://github.com/ | 1) The plugin can be downloaded from here [[https://github.com/aloctavodia/cheshift/archive/v3.6.zip Here]] | ||
2) If you are using the incentive version of PyMOL skip the next step and go directly to step 4. | |||
3) You should install NumPy and SciPy. These Python packages are available from the repositories of the main Linux distributions. Just use your default package manager (or command line) to install it. In Ubuntu/Debian you could do "sudo apt-get install python-numpy python-scipy". | |||
4) Open PyMOL and then go to plugin --> plugin manager --> Install New Plugin --> Install from local file --> Choose file. and choose the zip file you download in step 1. Now the plugin should be installed. | |||
If you have an old version of PyMOL without the plugin manager then, unzip the file downloaded in step 1 and copy the "cheshift" folder to the plugin directory, probably something like "/usr/lib/python2.7/dist-packages/pmg_tk/startup" | |||
=== Windows === | === Windows === | ||
This plugin have not been extensively tested on Windows machines, but it passed all the test I have done... | This plugin have not been extensively tested on Windows machines, but it passed all the test I have done... | ||
1) The plugin can be downloaded from here [[https://github.com/ | 1) The plugin can be downloaded from here [[https://github.com/aloctavodia/cheshift/archive/v3.6.zip Here]] | ||
2) You should install | 2) If you are using the incentive version of PyMOL skip the next step and go directly to step 4. | ||
3) You should install NumPy and SciPy. Probably the easiest way to do this on a Windows machine is to install a Python Distribution like [[https://store.continuum.io/cshop/anaconda/ Anaconda]] or [[https://www.enthought.com/products/canopy/ Canopy]] | |||
4) Open PyMOL and then go to plugin --> plugin manager --> Install New Plugin --> Install from local file --> Choose file. and choose the zip file you download in step 1. On Windows machine this step could take a couple of minutes, just wait for the confirmation message. Now the plugin should be installed. | |||
If you have an old version of PyMOL without the plugin manager then, unzip the file downloaded in step 1 and copy the "cheshift" folder to the plugin directory, probably something like "C:\Python27\Lib\site-packages\pmg_tk\startup\" | |||
=== Mac OsX === | === Mac OsX === | ||
This plugin have not been tested on | This plugin have not been extensively tested on Windows machines, but it passed all the test I have done... | ||
1) The plugin can be downloaded from here [[https://github.com/aloctavodia/cheshift/archive/v3.6.zip Here]] | |||
2) If you are using the incentive version of PyMOL skip the next step and go directly to step 4. | |||
3) You should install NumPy and SciPy. Probably the easiest way to do this on a Windows machine is to install a Python Distribution like [[https://store.continuum.io/cshop/anaconda/ Anaconda]] or [[https://www.enthought.com/products/canopy/ Canopy]] | |||
4) Open PyMOL and then go to plugin --> plugin manager --> Install New Plugin --> Install from local file --> Choose file. and choose the zip file you download in step 1. Now the plugin should be installed. | |||
If you have an old version of PyMOL without the plugin manager then, unzip the file downloaded in step 1 and copy the "cheshift" folder to the plugin directory, probably something like "/usr/lib/python2.7/dist-packages/pmg_tk/startup" | |||
== Using the Plugin for predicting <sup>13</sup>C<sup>α</sup> and <sup>13</sup>C<sup>β</sup> chemical shifts== | |||
1) Launch PyMOL and select a PDB file <br> | |||
2) Select "Cheshift" from the plugin menu. <br> | |||
3) Click the "Run" button.<br> | |||
4) The results will be saved in your working directory as a .txt file<br> | |||
== Using the Plugin == | == Using the Plugin for protein structure validation == | ||
1) Launch PyMOL and select | 1) Launch PyMOL and select a PDB file <br> | ||
2) Select | 2) Select "Cheshift" from the plugin menu. <br> | ||
3) Select a file with the experimental chemical shift values.<br> | 3) Select a file with the experimental chemical shift values.<br> | ||
4) Click the " | 4) Click the "Run" button.<br> | ||
5) Wait until results are displayed (this could take a few minutes, depending | 5) Wait until results are displayed (this could take from seconds to a few minutes, depending on the size of your protein and the speed of your computer<br> | ||
<b>Note:</b> | <b>Note:</b> | ||
* If the PDB file has more than one chain only the first one will be analysed. | |||
* If the PDB file has more than one chain only the first one will be | * The PDB file must have no missing residues. | ||
* The PDB file must | * Missing observed <sup>13</sup>C<sup>α</sup> and <sup>13</sup>C<sup>β</sup> chemical shifts are tolerated. | ||
* Missing observed <sup>13</sup>C<sup>α</sup> chemical shifts are tolerated. | * A file with observed <sup>13</sup>C<sup>α</sup> and/or <sup>13</sup>C<sup>β</sup> chemical shift values is needed. The format of this file should be the one used in the BMRB or the PDB. Alternatively, you can provide a file with the following format the first line should contain the name of the reference compound i.e DSS, TSP or TMS. The following lines should have four columns. The first column should be the residue number, the second the residue name (three-letter code) and the third column the <sup>13</sup>C<sup>α</sup> experimental chemical shifts and the last column the <sup>13</sup>C<sup>β</sup> chemical shifts. Spaces should be used to separates the columns. | ||
* | |||
DSS<br> | |||
1 MET 55.63 32.95<br> | |||
2 TYR 62.81 39.27<br> | |||
3 ALA 53.78 18.97<br> | |||
4 GLY 47.24 999.00<br> | |||
5 LYS 57.55 32.77<br> | |||
6 ILE 56.38 38.59<br> | |||
== License == | |||
<i>Che</i>Shift plugin is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License. A complete copy of the GNU General Public License can be accessed [http://www.gnu.org/licenses here]. | |||
<i>Che</i>Shift uses data derived from the [http://dunbrack.fccc.edu/ndrd/ Neighbor-Dependent Ramachandran Distributions] obtained by the Dunbrack Lab. This derived data is also released under the GPL license, with the permission of Professor Roland Dunbrack. The full NDRD data is released using a different [http://dunbrack.fccc.edu/ndrd/license/index.html license]. | |||
== | == Change log == | ||
< | |||
* 2014-07-02 (Version 3.6) | |||
Fix bug with relative path and add computation of the conformationally averaged rmsd (ca_rmsd). The ca_rmsd is the root mean square deviation of the theoretical chemical shift and the observed chemical shifts. The ca_rmsd is calculated for both nuclei (<sup>13</sup>C<sup>α</sup> and <sup>13</sup>C<sup>β</sup>). If more than one conformation is used, the theoretical chemical shift for each residue is the average along all conformations. | |||
* 2014-05-23 (Version 3.5) | |||
Previous versions required an Internet connection. This is the first stand alone version, i.e. all computations are performed on the local machine. | |||
This version of the plugin performs the same computations the <i>Che</i>Shift Server does. For details please read [1] | |||
<i>Che</i>Shift Server | * 2013-08-28 (Version 3.0) | ||
This version was able to establish a connection to the version of the <i>Che</i>Shift Server described in [1] | |||
* 2012-02-14 (Version 2.0) | |||
This version was able to establish a connection to the version of the <i>Che</i>Shift Server described in [2] | |||
== Citation == | == Citation == | ||
If you find this plugin useful please cite it as: | |||
Martin O.A. Vila J.A. and Scheraga H.A. (2012). <i>Che</i>Shift-2: Graphic validation of protein structures. Bioinformatics 2012. 28(11), 1538-1539. | |||
== References == | == References == | ||
Martin O.A. | [1] Martin O.A. Arnautova Y.A. Icazatti A.A. Scheraga H.A. and Vila J.A. A Physics-Based Method to Validate and Repair Flaws in Protein Structures. Proc Natl Acad Sci USA 2013, vol. 110 16826-16831. | ||
Vila J.A. Arnautova Y.A. Martin O.A. and Scheraga, H.A. (2009). Quantum-mechanics-derived | [2] Vila J.A. Arnautova Y.A. Martin O.A. and Scheraga, H.A. (2009). Quantum-mechanics-derived <sup>13</sup>C chemical shift server (<i>Che</i>Shift) for protein structure validation. PNAS, 106(40), 16972-16977. | ||
Vila, J.A. and Scheraga H.A. (2009). Assessing the accuracy of protein structures by quantum mechanical computations of <sup>13</sup>C<sup>α</sup> chemical shifts. Accounts of chemical research, 42(10), 1545-53. | [3] Vila, J.A. and Scheraga H.A. (2009). Assessing the accuracy of protein structures by quantum mechanical computations of <sup>13</sup>C<sup>α</sup> chemical shifts. Accounts of chemical research, 42(10), 1545-53. | ||
[[Category:Plugins]] | [[Category:Plugins]] | ||
[[Category:Pymol-script-repo]] | [[Category:Pymol-script-repo]] | ||
Latest revision as of 13:03, 16 January 2017
| Type | PyMOL Plugin |
|---|---|
| Download | https://github.com/aloctavodia/cheshift/archive/v3.6.zip |
| Author(s) | Osvaldo Martin |
| License | GPL |
Description
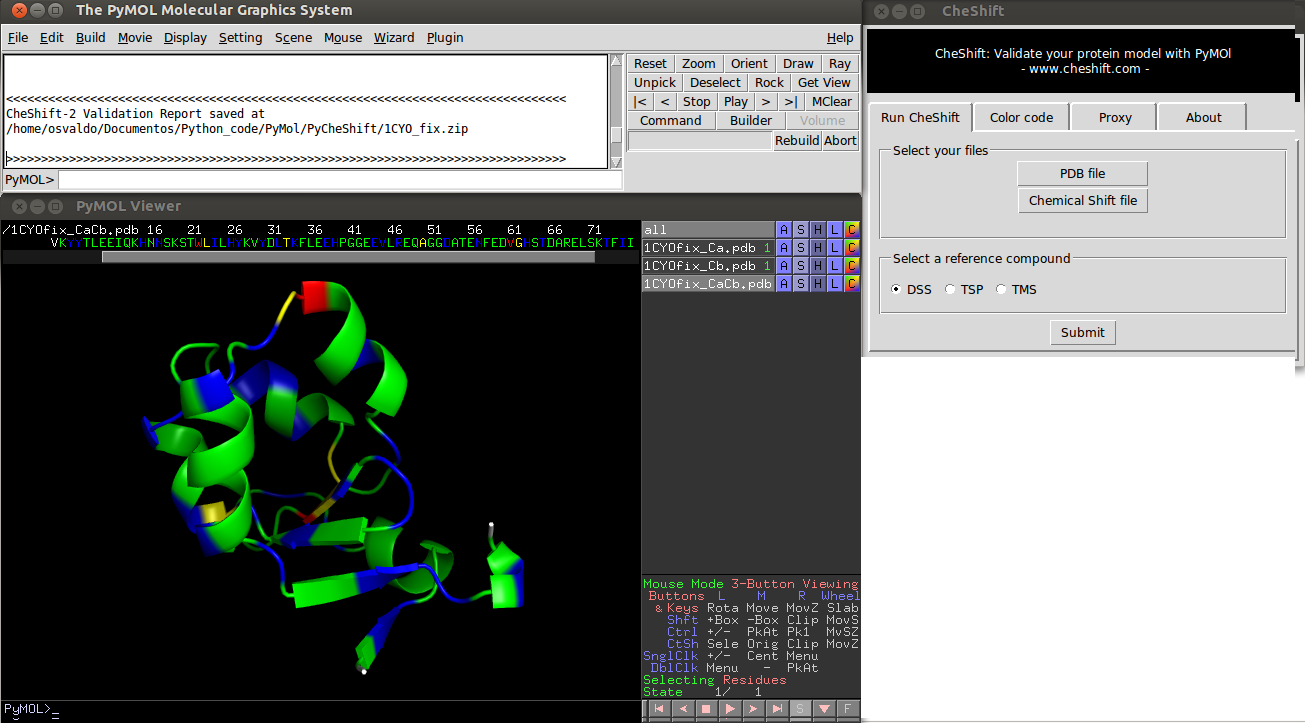
Colors indicate the difference between predicted and observed 13Cα and/or 13Cβchemical shifts values averaged over all uploaded conformers. Green, yellow and red colors represent small, medium and large differences, respectively. White is used if either the prediction failed or the observed value is missing.
Blue is used to highlight residues for which the agreement between observed and predicted 13Cα and 13Cβchemical shifts can be improved, i.e., if the (χ1 /χ2 ) side-chain
CheShift (pronounced /tʃeʃɪft/) is a software for prediction of 13Cα and 13Cβ chemical shifts and validation of proteins structures. It is based on the idea that the differences between observed and predicted 13Cα and 13Cβ chemical shifts can be used as a sensitive probe with which to detect possible local flaws in protein structures. A Web Server is also available.
This plugin provides a way to use PyMOL to validate a protein model using observed chemical shifts.
Version
The current version of this plugin is 3.6
Installation
Linux
1) The plugin can be downloaded from here [Here]
2) If you are using the incentive version of PyMOL skip the next step and go directly to step 4.
3) You should install NumPy and SciPy. These Python packages are available from the repositories of the main Linux distributions. Just use your default package manager (or command line) to install it. In Ubuntu/Debian you could do "sudo apt-get install python-numpy python-scipy".
4) Open PyMOL and then go to plugin --> plugin manager --> Install New Plugin --> Install from local file --> Choose file. and choose the zip file you download in step 1. Now the plugin should be installed. If you have an old version of PyMOL without the plugin manager then, unzip the file downloaded in step 1 and copy the "cheshift" folder to the plugin directory, probably something like "/usr/lib/python2.7/dist-packages/pmg_tk/startup"
Windows
This plugin have not been extensively tested on Windows machines, but it passed all the test I have done...
1) The plugin can be downloaded from here [Here]
2) If you are using the incentive version of PyMOL skip the next step and go directly to step 4.
3) You should install NumPy and SciPy. Probably the easiest way to do this on a Windows machine is to install a Python Distribution like [Anaconda] or [Canopy]
4) Open PyMOL and then go to plugin --> plugin manager --> Install New Plugin --> Install from local file --> Choose file. and choose the zip file you download in step 1. On Windows machine this step could take a couple of minutes, just wait for the confirmation message. Now the plugin should be installed. If you have an old version of PyMOL without the plugin manager then, unzip the file downloaded in step 1 and copy the "cheshift" folder to the plugin directory, probably something like "C:\Python27\Lib\site-packages\pmg_tk\startup\"
Mac OsX
This plugin have not been extensively tested on Windows machines, but it passed all the test I have done...
1) The plugin can be downloaded from here [Here]
2) If you are using the incentive version of PyMOL skip the next step and go directly to step 4.
3) You should install NumPy and SciPy. Probably the easiest way to do this on a Windows machine is to install a Python Distribution like [Anaconda] or [Canopy]
4) Open PyMOL and then go to plugin --> plugin manager --> Install New Plugin --> Install from local file --> Choose file. and choose the zip file you download in step 1. Now the plugin should be installed. If you have an old version of PyMOL without the plugin manager then, unzip the file downloaded in step 1 and copy the "cheshift" folder to the plugin directory, probably something like "/usr/lib/python2.7/dist-packages/pmg_tk/startup"
Using the Plugin for predicting 13Cα and 13Cβ chemical shifts
1) Launch PyMOL and select a PDB file
2) Select "Cheshift" from the plugin menu.
3) Click the "Run" button.
4) The results will be saved in your working directory as a .txt file
Using the Plugin for protein structure validation
1) Launch PyMOL and select a PDB file
2) Select "Cheshift" from the plugin menu.
3) Select a file with the experimental chemical shift values.
4) Click the "Run" button.
5) Wait until results are displayed (this could take from seconds to a few minutes, depending on the size of your protein and the speed of your computer
Note:
- If the PDB file has more than one chain only the first one will be analysed.
- The PDB file must have no missing residues.
- Missing observed 13Cα and 13Cβ chemical shifts are tolerated.
- A file with observed 13Cα and/or 13Cβ chemical shift values is needed. The format of this file should be the one used in the BMRB or the PDB. Alternatively, you can provide a file with the following format the first line should contain the name of the reference compound i.e DSS, TSP or TMS. The following lines should have four columns. The first column should be the residue number, the second the residue name (three-letter code) and the third column the 13Cα experimental chemical shifts and the last column the 13Cβ chemical shifts. Spaces should be used to separates the columns.
DSS
1 MET 55.63 32.95
2 TYR 62.81 39.27
3 ALA 53.78 18.97
4 GLY 47.24 999.00
5 LYS 57.55 32.77
6 ILE 56.38 38.59
License
CheShift plugin is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License. A complete copy of the GNU General Public License can be accessed here.
CheShift uses data derived from the Neighbor-Dependent Ramachandran Distributions obtained by the Dunbrack Lab. This derived data is also released under the GPL license, with the permission of Professor Roland Dunbrack. The full NDRD data is released using a different license.
Change log
- 2014-07-02 (Version 3.6)
Fix bug with relative path and add computation of the conformationally averaged rmsd (ca_rmsd). The ca_rmsd is the root mean square deviation of the theoretical chemical shift and the observed chemical shifts. The ca_rmsd is calculated for both nuclei (13Cα and 13Cβ). If more than one conformation is used, the theoretical chemical shift for each residue is the average along all conformations.
- 2014-05-23 (Version 3.5)
Previous versions required an Internet connection. This is the first stand alone version, i.e. all computations are performed on the local machine. This version of the plugin performs the same computations the CheShift Server does. For details please read [1]
- 2013-08-28 (Version 3.0)
This version was able to establish a connection to the version of the CheShift Server described in [1]
- 2012-02-14 (Version 2.0)
This version was able to establish a connection to the version of the CheShift Server described in [2]
Citation
If you find this plugin useful please cite it as:
Martin O.A. Vila J.A. and Scheraga H.A. (2012). CheShift-2: Graphic validation of protein structures. Bioinformatics 2012. 28(11), 1538-1539.
References
[1] Martin O.A. Arnautova Y.A. Icazatti A.A. Scheraga H.A. and Vila J.A. A Physics-Based Method to Validate and Repair Flaws in Protein Structures. Proc Natl Acad Sci USA 2013, vol. 110 16826-16831.
[2] Vila J.A. Arnautova Y.A. Martin O.A. and Scheraga, H.A. (2009). Quantum-mechanics-derived 13C chemical shift server (CheShift) for protein structure validation. PNAS, 106(40), 16972-16977.
[3] Vila, J.A. and Scheraga H.A. (2009). Assessing the accuracy of protein structures by quantum mechanical computations of 13Cα chemical shifts. Accounts of chemical research, 42(10), 1545-53.