Color by conservation: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
PedroLacerda (talk | contribs) No edit summary |
||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = script | |type = script | ||
|filename = color_by_conservation.py | |filename = scripts/color_by_conservation.py | ||
|author = [[User:Inchoate|Jason Vertrees]] | |author = [[User:Inchoate|Jason Vertrees]] | ||
|license = Free | |license = Free | ||
Latest revision as of 20:38, 22 June 2025
| Type | Python Script |
|---|---|
| Download | scripts/color_by_conservation.py |
| Author(s) | Jason Vertrees |
| License | Free |
| This code has been put under version control in the project Pymol-script-repo | |
Overview
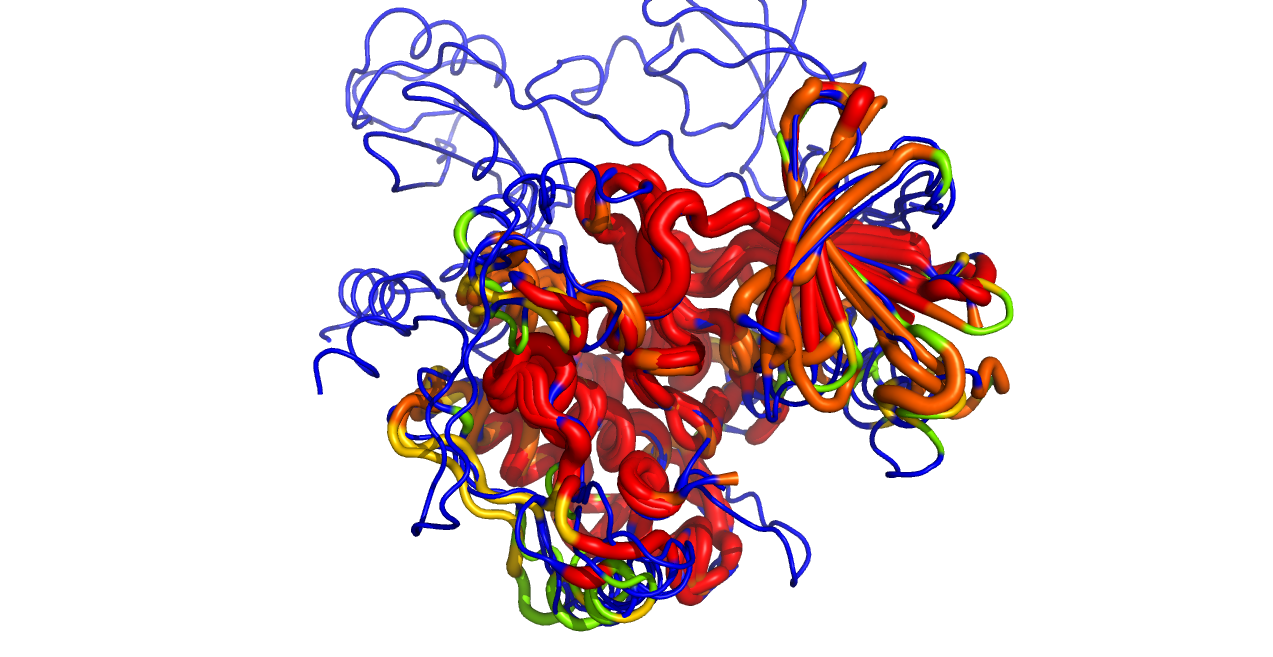
This script reads an alignment object and colors the protein objects in the alignment by the sequence conservation found in the alignment.
Example Usage
reinitialize
import color_by_conservation
# get some kinases
fetch 1opk 3dtc 3p86 2eva 3efw, async=0
# turn on the sequence viewer
set seq_view
# align them into the "algn" object
for x in cmd.get_names(): cmd.align(x, "3efw and c. A", object="algn")
# color
color_by_conservation aln=algn, as_putty=1