Colorbydisplacement: Difference between revisions
No edit summary |
PedroLacerda (talk | contribs) No edit summary |
||
| (2 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = script | |type = script | ||
|filename = colorbydisplacement.py | |filename = scripts/colorbydisplacement.py | ||
|author = [[ | |author = [[User:Tlinnet|Troels E. Linnet]] | ||
|license = BSD | |license = BSD | ||
}} | }} | ||
| Line 41: | Line 41: | ||
{{Template:PymolScriptRepoDownload|examples/colorbydisplacement_1.pml}} | {{Template:PymolScriptRepoDownload|examples/colorbydisplacement_1.pml}} | ||
<include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/colorbydisplacement_1.pml" highlight="python" /> | <include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/colorbydisplacement_1.pml" highlight="python" /> | ||
[[Category:Script_Library]] | [[Category:Script_Library]] | ||
[[Category:Structural_Biology_Scripts]] | [[Category:Structural_Biology_Scripts]] | ||
[[Category:Pymol-script-repo]] | |||
Latest revision as of 20:40, 22 June 2025
| Type | Python Script |
|---|---|
| Download | scripts/colorbydisplacement.py |
| Author(s) | Troels E. Linnet |
| License | BSD |
| This code has been put under version control in the project Pymol-script-repo | |
Introduction
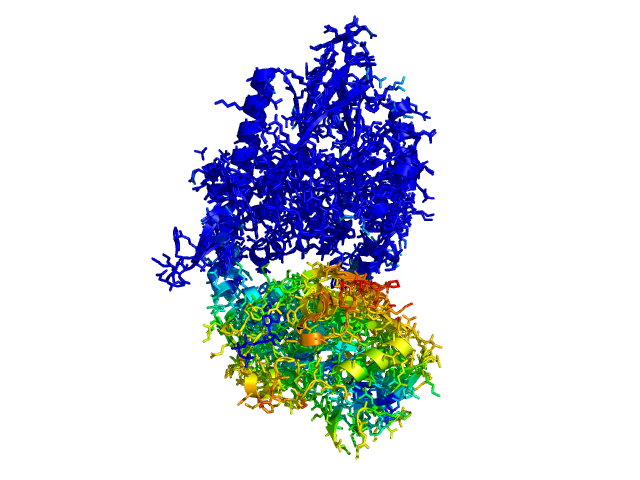
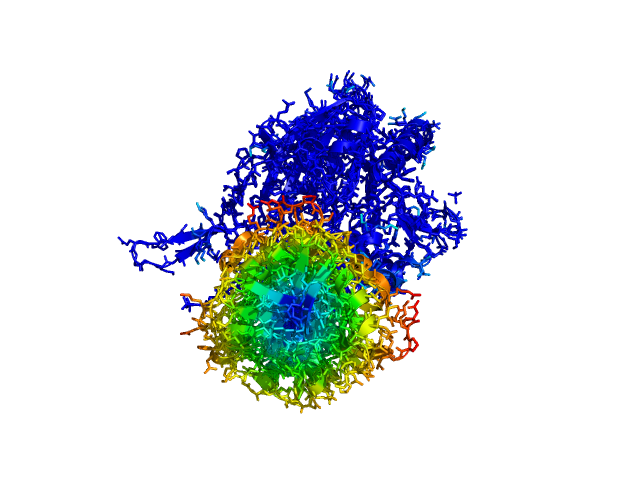
This script allows you to color two structures by distance displacement between an Open and Closed form of a protein, as calculated by PyMol's internal distance command. The pairwise distance is calculated between all-atoms. The distance displacement values are stored as B-factors of these residues, which are colored by a rainbow color spectrum, with blue specifying minimum and red indicating maximum.
Do keep in mind, all original B-factors values are overwritten!
There exist one version.
ColorByDisplacementAll is between All atoms in residues and is quite slow => 3-5 mins for a run. Ideal for sticks representation.
You have to specify which residues should be used in the alignment procedure, or it will take all residues as standard
V.2 is implemented the 2011.01.06 - Due to a bug in coloring.
Bug in code
A bug in the boolean operator of the spectrum command has been found. This versions work for version 1.3 Educational product.
For other versions of pymol, try to change (comment/uncomment) the cmd.spectrum line.
The other spectrum line works for Open-Source PyMOL 1.2r3pre, Incentive product
Examples
ColorByDisplacementAll O5NT, C5NT, super1=resi 26-355, super2=resi 26-355, doColor=t, doAlign=t
ColorByDisplacementAll O5NT, C5NT, super1=resi 26-355, super2=resi 26-355, doColor=t, doAlign=t, AlignedWhite='no'
Dark blue is low displacement, higher displacements are in orange/yellow/red.
Residues used for alignment is colored white. Can be turned off in top of algorithm.
Residues not in both pdb files is colored black
Example 1
| Download: examples/colorbydisplacement_1.pml | |
| This code has been put under version control in the project Pymol-script-repo | |
<include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/colorbydisplacement_1.pml" highlight="python" />