DynoPlot: Difference between revisions
(color support) |
(postscript dump) |
||
| Line 18: | Line 18: | ||
===USAGE=== | ===USAGE=== | ||
rama | |||
rama [ sel [, name [, symbols [, filename ]]]] | |||
===EXAMPLES=== | ===EXAMPLES=== | ||
<source lang="python"> | |||
fetch 1ENV, async=0 # (download it or use the PDB loader plugin) | |||
select sel01, resi 129-136 | |||
rama sel01 | |||
rock # the object needs to be moving in order for the angles to be updated. | |||
</source> | |||
Don't create callback object, use symbols by secondary structure and dump canvas as postscript file: | |||
<source lang="python"> | |||
fetch 2x19, async=0 | |||
color yellow, chain A | |||
color forest, chain B | |||
rama polymer, none, ss, /tmp/canvasdump.ps | |||
rama ss H, none, aa, /tmp/canvasdump_helix.ps | |||
rama ss S, none, aa, /tmp/canvasdump_sheet.ps | |||
</source> | |||
===SCRIPTS (DynoPlot.py)=== | ===SCRIPTS (DynoPlot.py)=== | ||
| Line 70: | Line 85: | ||
self.shapes = {} # store plot data, x,y etc.. | self.shapes = {} # store plot data, x,y etc.. | ||
self.idx2resn = {} # residue name mapping | self.idx2resn = {} # residue name mapping | ||
self.symbols = 0 # 0: amino acids, 1: secondary structure | |||
def axis(self,xmin=40,xmax=300,ymin=10,ymax=290,xint=290,yint=40,xlabels=[],ylabels=[]): | def axis(self,xmin=40,xmax=300,ymin=10,ymax=290,xint=290,yint=40,xlabels=[],ylabels=[]): | ||
| Line 115: | Line 131: | ||
resn, color, ss = self.idx2resn.get(meta) | resn, color, ss = self.idx2resn.get(meta) | ||
if | if self.symbols == 0: | ||
# symbols by amino acid (G/P/other) | # symbols by amino acid (G/P/other) | ||
mark = {'GLY': 'Tri', 'PRO': 'Rect'}.get(resn, 'Oval') | mark = {'GLY': 'Tri', 'PRO': 'Rect'}.get(resn, 'Oval') | ||
| Line 293: | Line 309: | ||
# New Callback object, so that we can update the structure when phi,psi points are moved. | # New Callback object, so that we can update the structure when phi,psi points are moved. | ||
class DynoRamaObject: | class DynoRamaObject: | ||
def __init__(self, selection=None, name=None): | def __init__(self, selection=None, name=None, symbols=''): | ||
from pymol import _ext_gui as pmgapp | from pymol import _ext_gui as pmgapp | ||
if pmgapp is not None: | if pmgapp is not None: | ||
| Line 314: | Line 330: | ||
ylabels=[-180,-150,-120,-90,-60,-30,0,30,60,90,120,150,180]) | ylabels=[-180,-150,-120,-90,-60,-30,0,30,60,90,120,150,180]) | ||
canvas.update() | canvas.update() | ||
if symbols == 'ss': | |||
canvas.symbols = 1 | |||
if name is None: | if name is None: | ||
| Line 357: | Line 376: | ||
if self.lock: | if self.lock: | ||
return | return | ||
# Loop through each item on plot to see if updated | # Loop through each item on plot to see if updated | ||
for value in self.canvas.shapes.itervalues(): | for value in self.canvas.shapes.itervalues(): | ||
| Line 367: | Line 387: | ||
value[2] = 0 | value[2] = 0 | ||
def rama(sel='(all)', name=None): | def rama(sel='(all)', name=None, symbols='aa', filename=None): | ||
''' | ''' | ||
DESCRIPTION | DESCRIPTION | ||
| Line 381: | Line 401: | ||
angles when canvas points are dragged, or 'none' to not create a callback | angles when canvas points are dragged, or 'none' to not create a callback | ||
object {default: DynoRamaObject} | object {default: DynoRamaObject} | ||
symbols = string: aa for amino acid or ss for secondary structure {default: aa} | |||
filename = string: filename for postscript dump of canvas {default: None} | |||
''' | ''' | ||
DynoRamaObject(sel, name) | dyno = DynoRamaObject(sel, name, symbols) | ||
if filename is not None: | |||
dyno.canvas.postscript(file=filename) | |||
# Extend these commands | # Extend these commands | ||
Revision as of 05:38, 18 November 2011
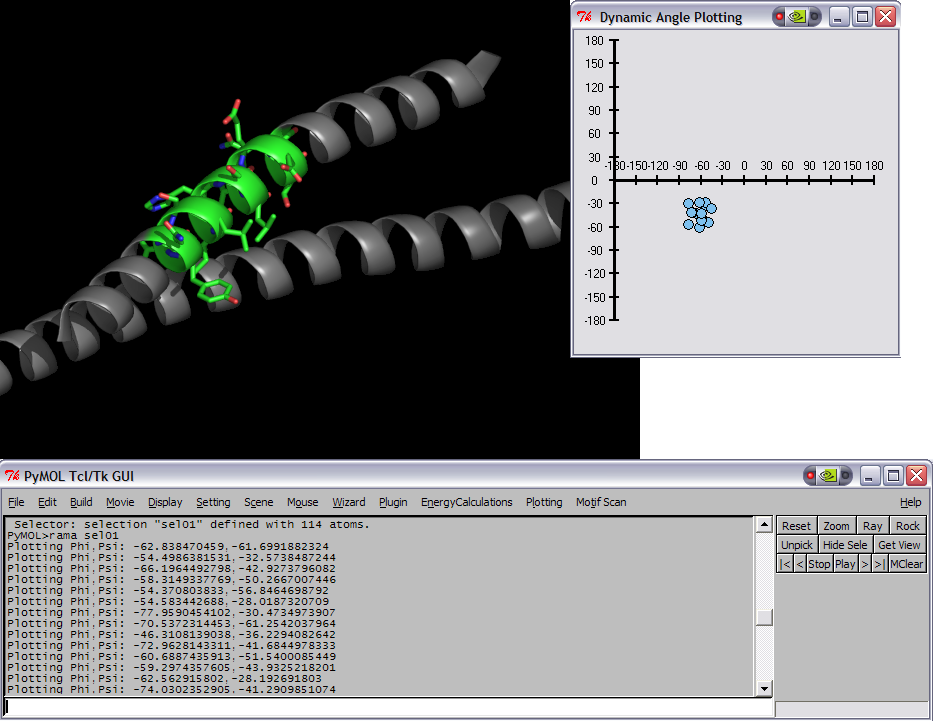
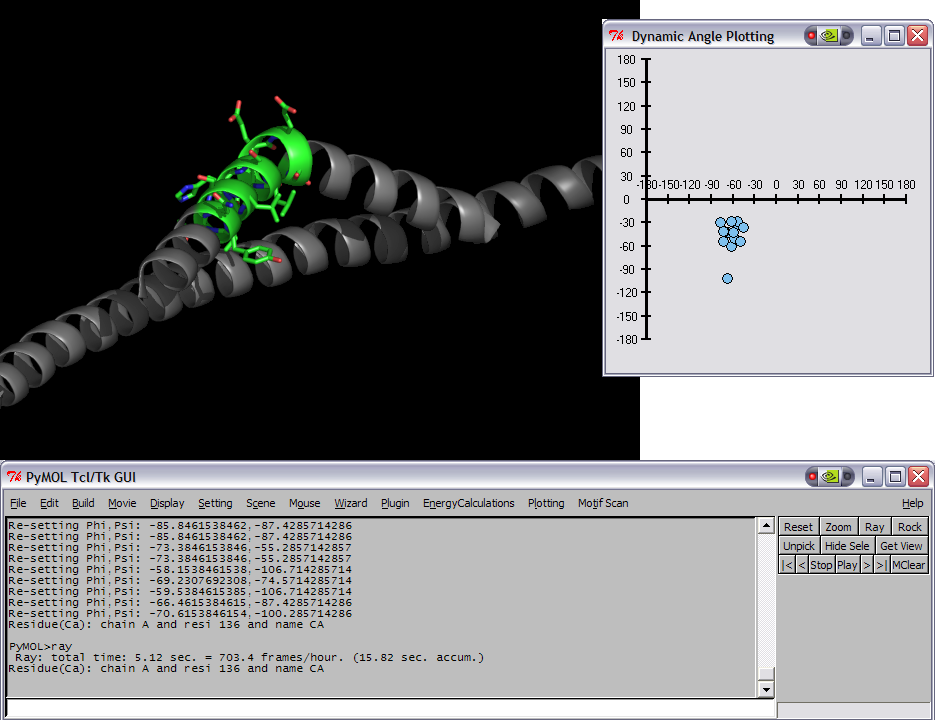
This script was setup to do generic plotting, that is given a set of data and axis labels it would create a plot. Initially, I had it setup to draw the plot directly in the PyMol window (allowing for both 2D and 3D style plots), but because I couldn't figure out how to billboard CGO objects (Warren told me at the time that it couldn't be done) I took a different approach. The plot now exists in it's own window and can only do 2D plots. It is however interactive. I only have here a Rama.(phi,psi) plot, but the code can be easily extended to other types of data. For instance, I had this working for an energy vs distance data that I had generated by another script.
This script will create a Phi vs Psi(Ramachandran) plot of the selection given. The plot will display data points which can be dragged around Phi,Psi space with the corresponding residue's Phi,Psi angles changing in the structure (PyMol window).
IMAGES
SETUP
Install from the plugins menu with Plugin > Manage Plugins > Install ... or just run the script.
NOTES / STATUS
- Tested on Linux, PyMol version 1.4
- Left, Right mouse buttons do different things; Right = identify data point, Left = drag data point around
- Post comments/questions or send them to: dwkulp@mail.med.upenn.edu
USAGE
rama [ sel [, name [, symbols [, filename ]]]]
EXAMPLES
fetch 1ENV, async=0 # (download it or use the PDB loader plugin)
select sel01, resi 129-136
rama sel01
rock # the object needs to be moving in order for the angles to be updated.
Don't create callback object, use symbols by secondary structure and dump canvas as postscript file:
fetch 2x19, async=0
color yellow, chain A
color forest, chain B
rama polymer, none, ss, /tmp/canvasdump.ps
rama ss H, none, aa, /tmp/canvasdump_helix.ps
rama ss S, none, aa, /tmp/canvasdump_sheet.ps
SCRIPTS (DynoPlot.py)
DynoPlot.py
###############################################
# File: DynoPlot.py
# Author: Dan Kulp
# Creation Date: 8/29/05
#
# Modified 2011-11-17 by Thomas Holder
#
# Notes:
# Draw plots that display interactive data.
# Phi,Psi plot shown.
###############################################
from __future__ import division
from __future__ import generators
import Tkinter
from pymol import cmd
# workaround: Set to True if nothing gets drawn on canvas, for example on linux with "pymol -x"
with_mainloop = False
class SimplePlot(Tkinter.Canvas):
# Class variables
mark_size = 4
def __init__(self, *args, **kwargs):
Tkinter.Canvas.__init__(self, *args, **kwargs)
self.xlabels = [] # axis labels
self.ylabels = []
self.spacingx = 0 # spacing in x direction
self.spacingy = 0
self.xmin = 0 # min value from each axis
self.ymin = 0
self.lastx = 0 # previous x,y pos of mouse
self.lasty = 0
self.isdown = 0 # flag for mouse pressed
self.item = (0,) # items array used for clickable events
self.shapes = {} # store plot data, x,y etc..
self.idx2resn = {} # residue name mapping
self.symbols = 0 # 0: amino acids, 1: secondary structure
def axis(self,xmin=40,xmax=300,ymin=10,ymax=290,xint=290,yint=40,xlabels=[],ylabels=[]):
# Store variables in self object
self.xlabels = xlabels
self.ylabels = ylabels
self.spacingx = (xmax-xmin) / (len(xlabels) - 1)
self.spacingy = (ymax-ymin) / (len(ylabels) - 1)
self.xmin = xmin
self.ymin = ymin
# Create axis lines
self.create_line((xmin,xint,xmax,xint),fill="black",width=3)
self.create_line((yint,ymin,yint,ymax),fill="black",width=3)
# Create tick marks and labels
nextspot = xmin
for label in xlabels:
self.create_line((nextspot, xint+5,nextspot, xint-5),fill="black",width=2)
self.create_text(nextspot, xint-15, text=label)
if len(xlabels) == 1:
nextspot = xmax
else:
nextspot += (xmax - xmin)/ (len(xlabels) - 1)
nextspot = ymax
for label in ylabels:
self.create_line((yint+5,nextspot,yint-5,nextspot),fill="black",width=2)
self.create_text(yint-20,nextspot,text=label)
if len(ylabels) == 1:
nextspot = ymin
else:
nextspot -= (ymax - ymin)/ (len(ylabels) - 1)
# Plot a point
def plot(self,xp,yp,meta):
# Convert from 'label' space to 'pixel' space
x = self.convertToPixel("X",xp)
y = self.convertToPixel("Y",yp)
resn, color, ss = self.idx2resn.get(meta)
if self.symbols == 0:
# symbols by amino acid (G/P/other)
mark = {'GLY': 'Tri', 'PRO': 'Rect'}.get(resn, 'Oval')
else:
# symbols by secondary structure
mark = {'H': 'Oval', 'S': 'Rect'}.get(ss, 'Tri')
if mark == 'Oval':
create_shape = self.create_oval
coords = [x-self.mark_size, y-self.mark_size,
x+self.mark_size, y+self.mark_size]
elif mark == 'Tri':
create_shape = self.create_polygon
coords = [x, y-self.mark_size,
x+self.mark_size, y+self.mark_size,
x-self.mark_size, y+self.mark_size]
else:
create_shape = self.create_rectangle
coords = [x-self.mark_size, y-self.mark_size,
x+self.mark_size, y+self.mark_size]
if color >= 0x40000000:
color = '#%06x' % (color & 0xffffff)
else:
color = '#%02x%02x%02x' % tuple([255*i
for i in cmd.get_color_tuple(color)])
oval = create_shape(*coords,
width=1, outline="black", fill=color)
self.shapes[oval] = [x,y,0,xp,yp,meta]
# Convert from pixel space to label space
def convertToLabel(self,axis, value):
# Defaultly use X-axis info
label0 = self.xlabels[0]
label1 = self.xlabels[1]
spacing = self.spacingx
min = self.xmin
# Set info for Y-axis use
if axis == "Y":
label0 = self.ylabels[0]
label1 = self.ylabels[1]
spacing = self.spacingy
min = self.ymin
pixel = value - min
label = pixel / spacing
label = label0 + label * abs(label1 - label0)
if axis == "Y":
label = - label
return label
# Converts value from 'label' space to 'pixel' space
def convertToPixel(self,axis, value):
# Defaultly use X-axis info
label0 = self.xlabels[0]
label1 = self.xlabels[1]
spacing = self.spacingx
min = self.xmin
# Set info for Y-axis use
if axis == "Y":
label0 = self.ylabels[0]
label1 = self.ylabels[1]
spacing = self.spacingy
min = self.ymin
# Get axis increment in 'label' space
inc = abs(label1 - label0)
# 'Label' difference from value and smallest label (label0)
diff = float(value - label0)
# Get whole number in 'label' space
whole = int(diff / inc)
# Get fraction number in 'label' space
part = float(float(diff/inc) - whole)
# Return 'pixel' position value
pixel = whole * spacing + part * spacing
# Reverse number by subtracting total number of pixels - value pixels
if axis == "Y":
tot_label_diff = float(self.ylabels[-1] - label0)
tot_label_whole = int(tot_label_diff / inc)
tot_label_part = float(float(tot_label_diff / inc) - tot_label_whole)
tot_label_pix = tot_label_whole * spacing + tot_label_part *spacing
pixel = tot_label_pix - pixel
# Add min edge pixels
pixel = pixel + min
return pixel
# Print out which data point you just clicked on..
def pickWhich(self,event):
# Find closest data point
x = event.widget.canvasx(event.x)
y = event.widget.canvasx(event.y)
spot = event.widget.find_closest(x,y)
# Print the shape's meta information corresponding with the shape that was picked
if spot[0] in self.shapes:
cmd.select('sele', '(%s`%d)' % self.shapes[spot[0]][5])
cmd.iterate('sele', 'print " You clicked /%s/%s/%s/%s`%s/%s (DynoPlot)" %' + \
' (model, segi, chain, resn, resi, name)')
cmd.center('byres sele', animate=1)
# Mouse Down Event
def down(self,event):
# Store x,y position
self.lastx = event.x
self.lasty = event.y
# Find the currently selected item
x = event.widget.canvasx(event.x)
y = event.widget.canvasx(event.y)
self.item = event.widget.find_closest(x,y)
# Identify that the mouse is down
self.isdown = 1
# Mouse Up Event
def up(self,event):
# Get label space version of x,y
labelx = self.convertToLabel("X",event.x)
labely = self.convertToLabel("Y",event.y)
# Convert new position into label space..
if self.item[0] in self.shapes:
self.shapes[self.item[0]][0] = event.x
self.shapes[self.item[0]][1] = event.y
self.shapes[self.item[0]][2] = 1
self.shapes[self.item[0]][3] = labelx
self.shapes[self.item[0]][4] = labely
# Reset Flags
self.item = (0,)
self.isdown = 0
# Mouse Drag(Move) Event
def drag(self,event):
# Check that mouse is down and item clicked is a valid data point
if self.isdown and self.item[0] in self.shapes:
self.move(self.item, event.x - self.lastx, event.y - self.lasty)
self.lastx = event.x
self.lasty = event.y
def set_phipsi(model, index, phi, psi):
atsele = [
'first ((%s`%d) extend 2 and name C)' % (model, index), # prev C
'first ((%s`%d) extend 1 and name N)' % (model, index), # this N
'(%s`%d)' % (model, index), # this CA
'last ((%s`%d) extend 1 and name C)' % (model, index), # this C
'last ((%s`%d) extend 2 and name N)' % (model, index), # next N
]
try:
cmd.set_dihedral(atsele[0], atsele[1], atsele[2], atsele[3], phi)
cmd.set_dihedral(atsele[1], atsele[2], atsele[3], atsele[4], psi)
except:
print ' DynoPlot Error: cmd.set_dihedral failed'
# New Callback object, so that we can update the structure when phi,psi points are moved.
class DynoRamaObject:
def __init__(self, selection=None, name=None, symbols=''):
from pymol import _ext_gui as pmgapp
if pmgapp is not None:
import Pmw
rootframe = Pmw.MegaToplevel(pmgapp.root)
parent = rootframe.interior()
else:
rootframe = Tkinter.Tk()
parent = rootframe
rootframe.title(' Dynamic Angle Plotting ')
rootframe.protocol("WM_DELETE_WINDOW", self.close_callback)
canvas = SimplePlot(parent,width=320,height=320)
canvas.bind("<Button-2>",canvas.pickWhich)
canvas.bind("<Button-3>",canvas.pickWhich)
canvas.pack(side=Tkinter.LEFT,fill="both",expand=1)
canvas.axis(xint=150,
xlabels=[-180,-120,-60,0,60,120,180],
ylabels=[-180,-150,-120,-90,-60,-30,0,30,60,90,120,150,180])
canvas.update()
if symbols == 'ss':
canvas.symbols = 1
if name is None:
try:
name = cmd.get_unused_name('DynoRama')
except AttributeError:
name = 'DynoRamaObject'
self.rootframe = rootframe
self.canvas = canvas
self.name = name
self.lock = 0
if name != 'none':
auto_zoom = cmd.get('auto_zoom')
cmd.set('auto_zoom', 0)
cmd.load_callback(self, name)
cmd.set('auto_zoom', auto_zoom)
canvas.bind("<ButtonPress-1>",canvas.down)
canvas.bind("<ButtonRelease-1>",canvas.up)
canvas.bind("<Motion>",canvas.drag)
if selection is not None:
self.start(selection)
if with_mainloop and pmgapp is None:
rootframe.mainloop()
def close_callback(self):
cmd.delete(self.name)
self.rootframe.destroy()
def start(self,sel):
self.lock = 1
cmd.iterate('(%s) and name CA' % sel,'idx2resn[model,index] = (resn, color, ss)',
space={'idx2resn': self.canvas.idx2resn})
for model_index, (phi,psi) in cmd.get_phipsi(sel).iteritems():
print " Plotting Phi,Psi: %8.2f,%8.2f" % (phi, psi)
self.canvas.plot(phi, psi, model_index)
self.lock = 0
def __call__(self):
if self.lock:
return
# Loop through each item on plot to see if updated
for value in self.canvas.shapes.itervalues():
# Look for update flag...
if value[2]:
# Set residue's phi,psi to new values
model, index = value[5]
print " Re-setting Phi,Psi: %8.2f,%8.2f" % (value[3],value[4])
set_phipsi(model, index, value[3], value[4])
value[2] = 0
def rama(sel='(all)', name=None, symbols='aa', filename=None):
'''
DESCRIPTION
Ramachandran Plot
http://pymolwiki.org/index.php/DynoPlot
ARGUMENTS
sel = string: atom selection {default: all}
name = string: name of callback object which is responsible for setting
angles when canvas points are dragged, or 'none' to not create a callback
object {default: DynoRamaObject}
symbols = string: aa for amino acid or ss for secondary structure {default: aa}
filename = string: filename for postscript dump of canvas {default: None}
'''
dyno = DynoRamaObject(sel, name, symbols)
if filename is not None:
dyno.canvas.postscript(file=filename)
# Extend these commands
cmd.extend('ramachandran', rama)
cmd.auto_arg[0]['ramachandran'] = cmd.auto_arg[0]['zoom']
# Add to plugin menu
def __init_plugin__(self):
self.menuBar.addcascademenu('Plugin', 'PlotTools', 'Plot Tools', label='Plot Tools')
self.menuBar.addmenuitem('PlotTools', 'command', 'Launch Rama Plot', label='Rama Plot',
command = lambda: DynoRamaObject('(enabled)'))
# vi:expandtab:smarttab