Propka: Difference between revisions
| Line 239: | Line 239: | ||
assert method in ['upload', 'file'], "'method' has to be either: method=upload or method=file" | assert method in ['upload', 'file'], "'method' has to be either: method=upload or method=file" | ||
assert molecule not in ['NIL'], "You always have to provide molecule name. Example: molecule=4ins" | assert molecule not in ['NIL'], "You always have to provide molecule name. Example: molecule=4ins" | ||
### If molecule="all", then try to get the first molecule | |||
if molecule=="all": | |||
assert len(cmd.get_names())!=0, "Did you forget to load a molecule? There are no objects in pymol." | |||
molecule=cmd.get_names()[0] | |||
### To print out to screen for selected residues. Can be separated with "." or make ranges with "-". Example: resi="4-8.10" | ### To print out to screen for selected residues. Can be separated with "." or make ranges with "-". Example: resi="4-8.10" | ||
if resi != "0": resi_range = ResiRange(resi) | if resi != "0": resi_range = ResiRange(resi) | ||
Revision as of 13:32, 16 August 2011
Acknowledgement
The PROPKA method is developed by the Jensen Research Group , Department of Chemistry, University of Copenhagen.
propka.py contact and relies on the result from the propka server
Introduction
This script can fetch the pka values for a protein from the propka server. The "propka" function downloads the results and processes them.
It also automatically writes a pymol command file and let pymol execute it. This command file make pka atoms, rename them, label them and color them according to the pka value.
If is not possible to get the mechanize module to work, one can go to the propka server, upload a .pdb file, and download the .pka file.
By setting: "method=file" and point to the .pka file "file_name=.\Results_propka\4ins_.pka", one can process the .pka result file and get the pymol command file.
The last possibility, is just to ask for the pka values of a recognized PDB id. This is done with the "getpropka" function.
Dependency of python module: mechanize
The script needs mechanize to run.
- On windows, it is not easy to make additional modules available for pymol. So put in into your working folder.
- The easy manual way:
- Go to: http://wwwsearch.sourceforge.net/mechanize/download.html
- Download mechanize-0.2.5.zip. http://pypi.python.org/packages/source/m/mechanize/mechanize-0.2.5.zip
- Extract to .\mechanize-0.2.5 then move the in-side folder "mechanize" to your folder with propka.py. The rest of .\mechanize-0.2.5 you don't need.
- You can also see other places where you could put the "mechanize" folder. Write this in pymol to see the paths where pymol is searching for "mechanize"
- import sys; print(sys.path)
Examples
### Point to your directory with the script.
#cd /homes/linnet/Documents/Speciale/5NT-project/Mutant-construct/predict_reactivity/propka
cd C:\Users\tlinnet\Documents\My Dropbox\Speciale\5NT-project\Mutant-construct\predict_reactivity\propka
run ./propka.py
fetch 4ins, async=0
propka OR
propka 4ins OR
propka molecule=4ins, chain=*, resi=19.20, resn=ASP.TYR, logtime=_, verbose=yes, showresult=no
fetch 1hp1, async=0
propka molecule=1hp1, chain=A, resi=308.513, resn=CYS, logtime=_, verbose=no, showresult=no
getpropka source=ID, PDBID=4ins, logtime=_, server_wait=3.0, verbose=yes, showresult=no
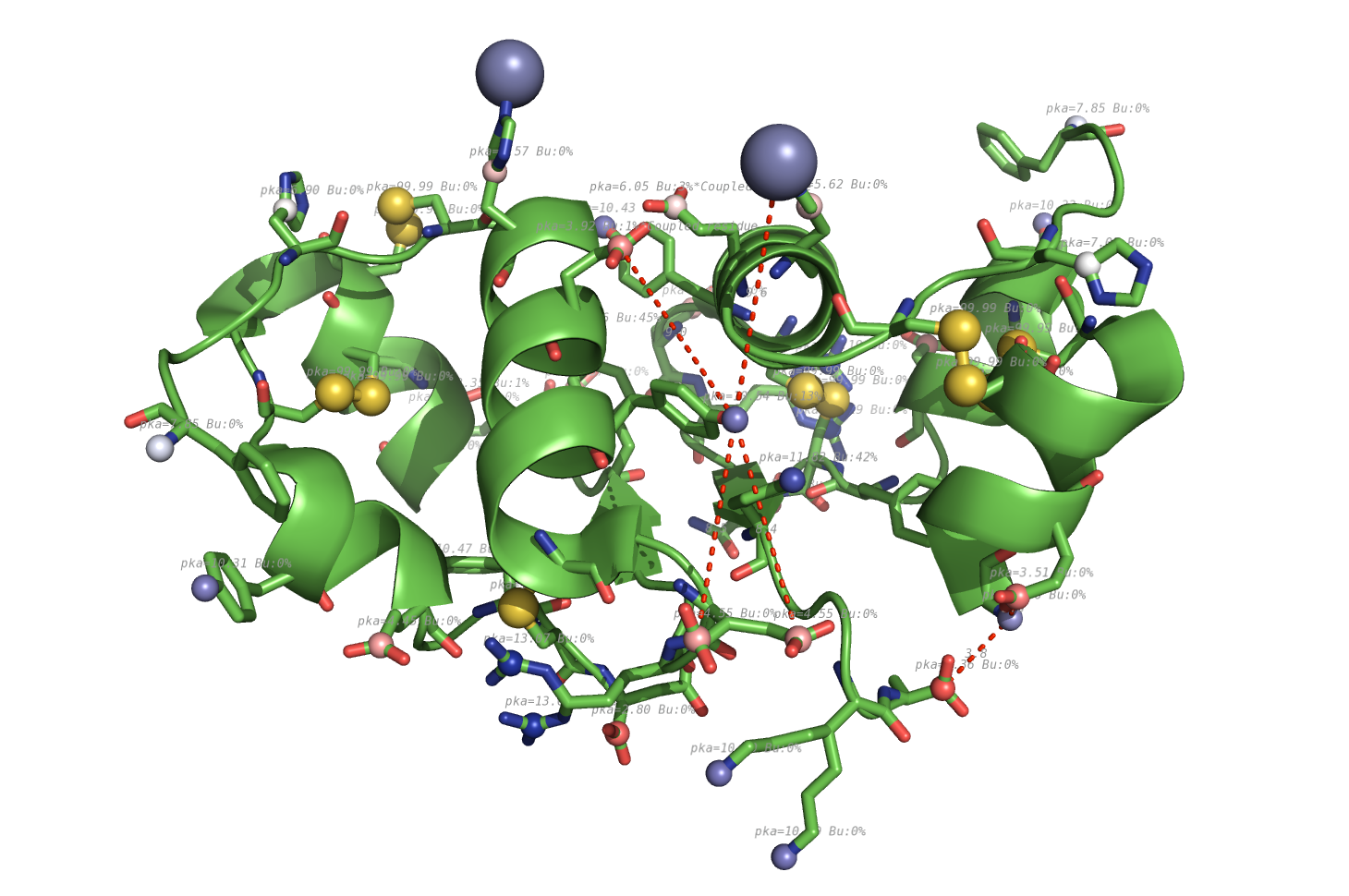
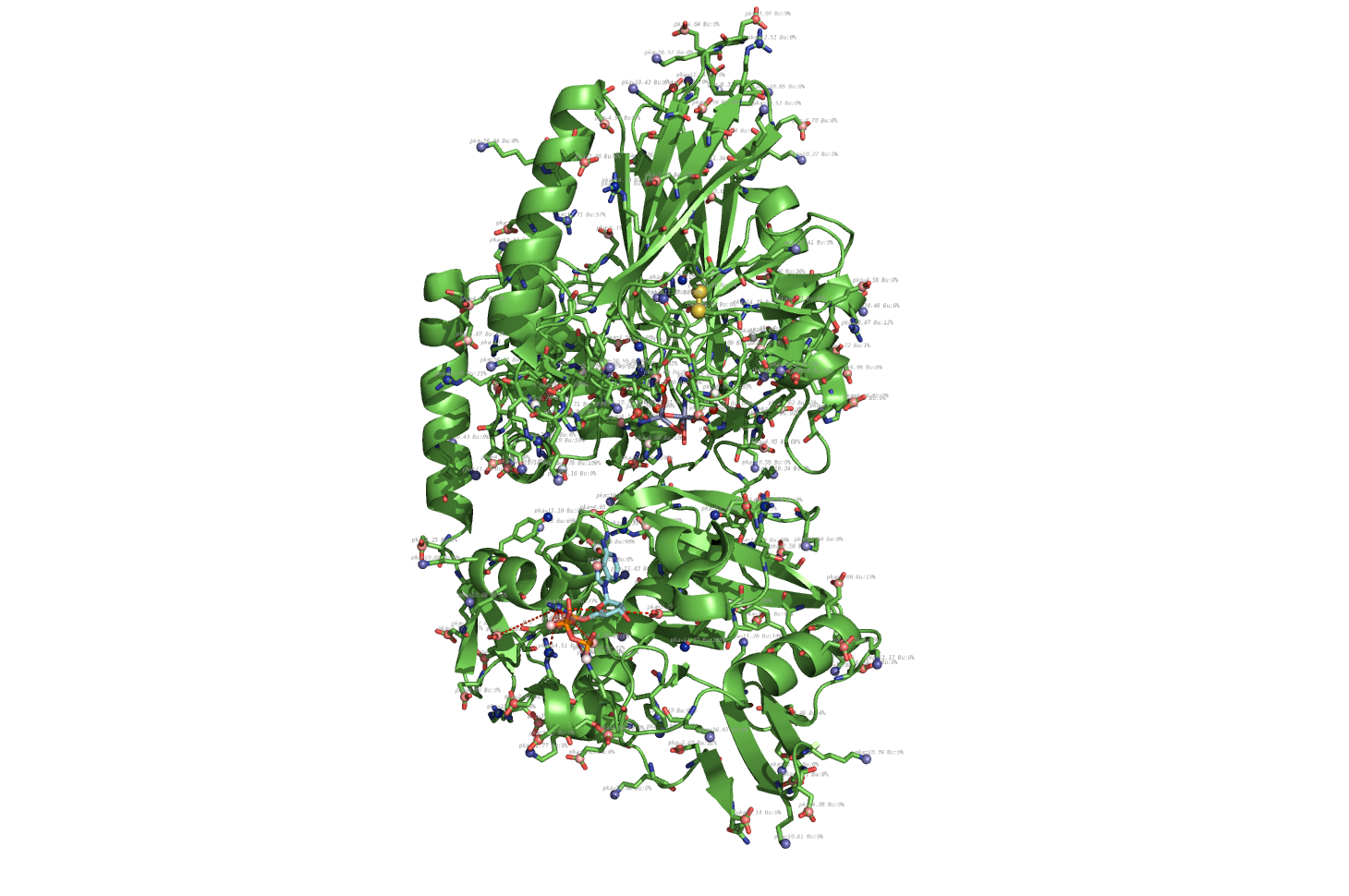
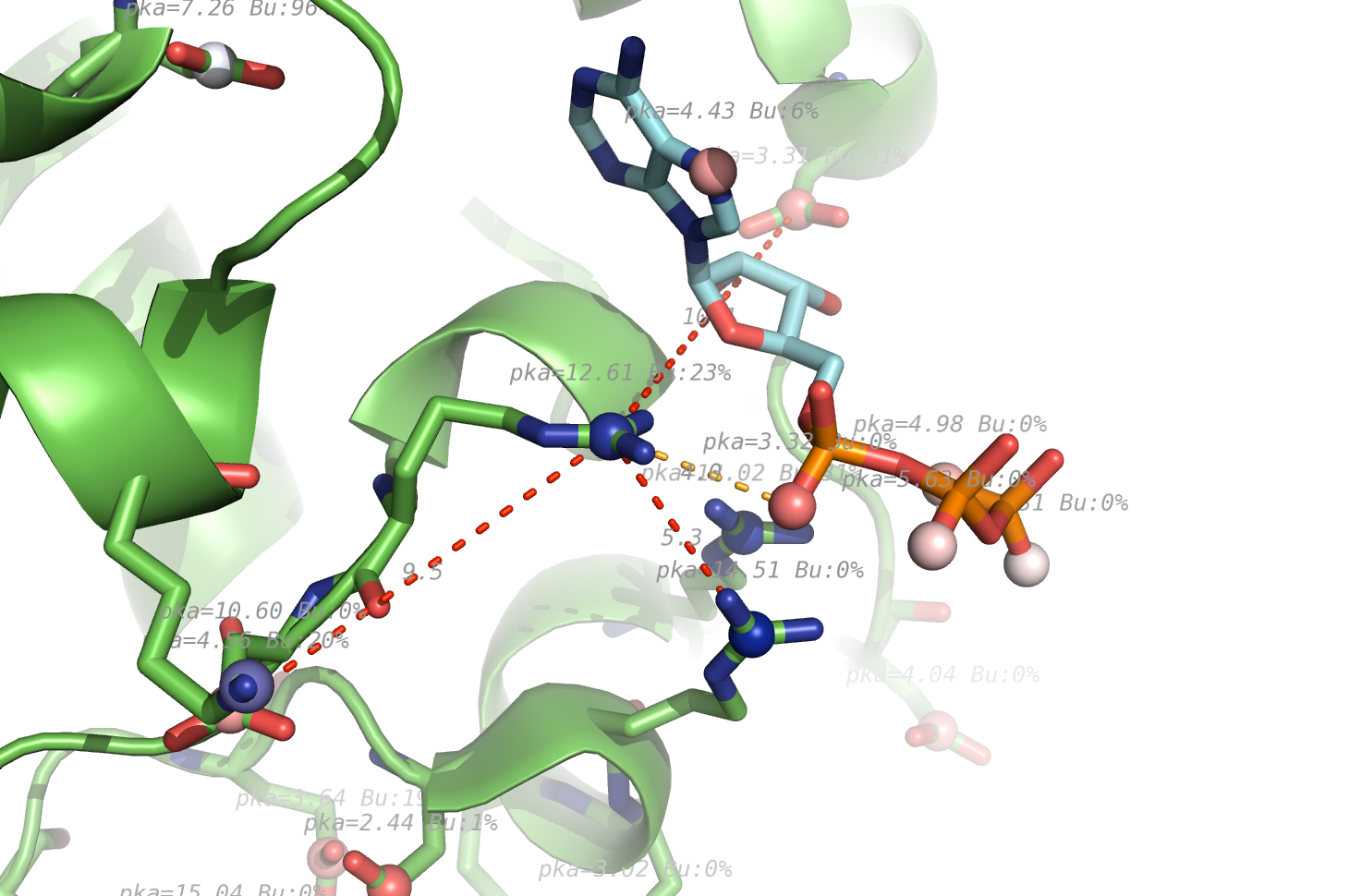
pka atoms are created and renamed for their pka value. That makes it easy to "click" the atom in pymol and instantly see the pka value.
The atoms b value are also altered to the pka value, and the atoms are then spectrum colored from pka=0-14.
The pka value of 99.9 represent a di-sulphide bond, and is colored gold and the sphere size is set a little bigger.
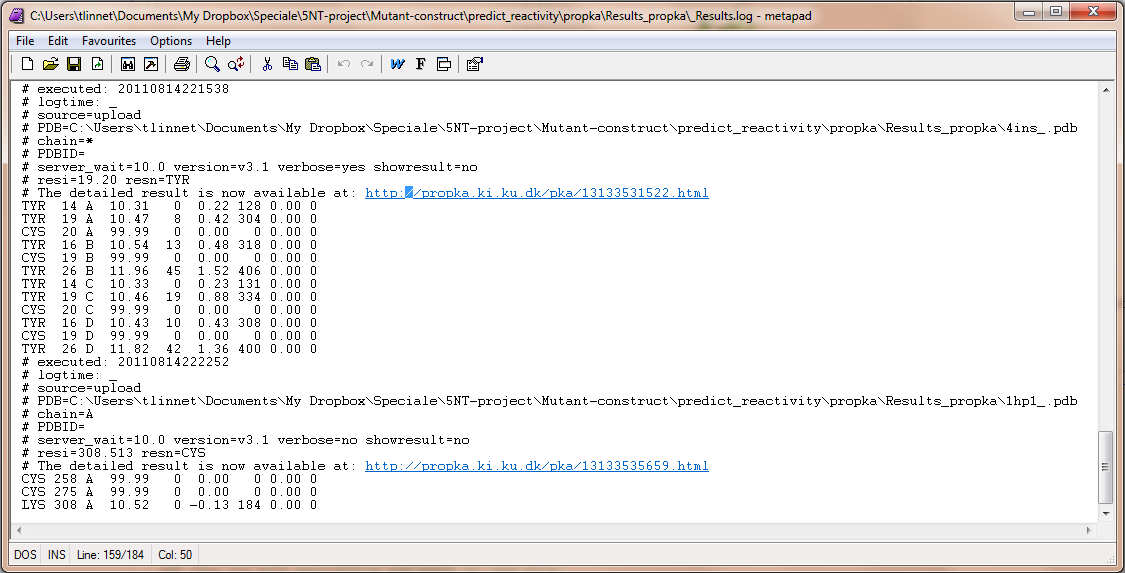
If one wants to see the specified result, the logfile ./Results_propka/_Results.log saves the link to the propka server. Here one can see in an interactive Jmol appp, the interactions to the pka residues.
Example Pymol Script
### Point to your directory with the script.
#cd /homes/linnet/Documents/Speciale/5NT-project/Mutant-construct/predict_reactivity/propka
cd C:\Users\tlinnet\Documents\My Dropbox\Speciale\5NT-project\Mutant-construct\predict_reactivity\propka
### If you have the script in your working directory the
run ./propka.py
### You can also make the script general available. Put it into your python path. Ex: C:\Program Files (x86)\PyMOL\PyMOL\modules Then do instead:
#import propka
### The fastest method is just to write propka. Then the pymol "all" molecule is assumed and send to server. But best to write molecule name
fetch 4ins, async=0
propka
propka 4ins
### Fetch 4ins from web. async make sure, we wait to have completed
#fetch 4ins, async=0
#propka molecule=4ins, chain=*, resi=19.20, resn=ASP.TYR, logtime=_, verbose=yes, showresult=no
### If there is problems to get the mechanize module working, get the result .pka file at the propka webpage manually.
### Then run with method=file and point to .pka file.
#fetch 4ins, async=0
#hide everything, 4ins
#show cartoon, 4ins
#propka method=file, molecule=4ins, resi=18.25-30, resn=cys, file_name=.\Results_propka\4ins_.pka
### Some more examples. This molecule has 550 residues, so takes a longer time.
#fetch 1hp1, async=0
#propka molecule=1hp1, chain=A, resi=308.513, resn=CYS, logtime=_, verbose=no, showresult=no
### One can also just make a lookup for a protein. Use function: getpropka
### Note. This does only print the result to the pymol command line
#getpropka source=ID, PDBID=4ake, logtime=_, server_wait=3.0, verbose=no, showresult=yes
#getpropka source=ID, PDBID=4ins, logtime=_, server_wait=3.0, verbose=yes, showresult=no
Input paramaters
# method : method=upload is default. This sends .pdb file and request result from propka server.
## method=file will only process a manual .pka file, and write a pymol command file. No use of mechanize.
# molecule : name of the molecule. Ending of file is assumed to be .pdb
# chain : which chains are saved to file, before molecule file is send to server. Separate with "." Ex: chain=A.b
# resi : Select by residue number, which residues should be printed to screen and saved to the log file: \Results_propka\_Results.log.
## Separate with "." or make ranges with "-". Ex: resi=35.40-50
# resn : Select by residue name, which residues should be printed to screen and saved to the log file: \Results_propka\_Results.log.
## Separate with "." Ex: resn=cys.tyr
# logtime : Each execution give a set of files with the job id=logtime. If logtime is not provided, the current time is used.
## Normal it usefull to set it empty. Ex: logtime=_
# verbose : Verbose is switch, to turn on messages for the mechanize section. This is handsome to see how mechanize works, and for error searching.
# showresult : Switch, to turn on all results in pymol command window. Ex: showresult=yes
# file_name: only used for: propka method=file . Write the path to .pka file
# server_wait=10.0 is default. This defines how long time between asking the server for a result. Set no lower than 3 seconds.
# version=v3.1 is default. This is what version of propka which would be used.
## Possible: 'v3.1','v3.0','v2.0'. If a newer version is available than the current v3.1, a error message is raised to make user update the script.
# source : source=upload is default and is set at the propka webpage. Can only be used for in the function "getpropka".
# getpropka source=ID, PDBID=4ake , one can print to the command line, the pka value for any official pdb ID. No files are displayed in pymol.
Python Code
The code can be downloaded fast from here http://tinyurl.com/pymolpropka.
- wget http://tinyurl.com/pymolpropka
- mv pymolpropka propka.py
#-------------------------------------------------------------------------------
# Name: propka for pymol
# Purpose: To fetch and display the pka values for protein of intetest
#
# Author: Troels E. Linnet
#
# Created: 14/08/2011
# Copyright: (c) Troels E. Linnet 2011
# Contact: tlinnet snabela gmail dot com
# Licence: Free for all
#
# Download: http://tinyurl.com/pymolpropka
#
#-------------------------------------------------------------------------------
"""
The PROPKA method is developed by the
Jensen Research Group
Department of Chemistry
University of Copenhagen
Please cite these references in publications:
Hui Li, Andrew D. Robertson, and Jan H. Jensen
"Very Fast Empirical Prediction and Interpretation of Protein pKa Values"
Proteins, 2005, 61, 704-721.
Delphine C. Bas, David M. Rogers, and Jan H. Jensen
"Very Fast Prediction and Rationalization of pKa Values for Protein-Ligand Complexes"
Proteins, 2008, 73, 765-783.
Mats H.M. Olsson, Chresten R. Soendergard, Michal Rostkowski, and Jan H. Jensen
"PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa predictions"
Journal of Chemical Theory and Computation, 2011 7 (2), 525-537
Chresten R. Soendergaard, Mats H.M. Olsson, Michaz Rostkowski, and Jan H. Jensen
"Improved Treatment of Ligands and Coupling Effects in Empirical Calculation and Rationalization of pKa Values"
Journal of Chemical Theory and Computation, 2011 in press
"""
#-------------------------------------------------------------------------------
# The script needs mechanize to run.
# On windows, it is not easy to make additional modules available for pymol. So put in into your working folder.
#1)The easy manual way:
#a)Go to: http://wwwsearch.sourceforge.net/mechanize/download.html
#b)Download mechanize-0.2.5.zip. http://pypi.python.org/packages/source/m/mechanize/mechanize-0.2.5.zip
#c)Extract to .\mechanize-0.2.5 then move the in-side folder "mechanize" to your folder with propka.py. The rest of .\mechanize-0.2.5 you don't need.
#You can also see other places where you could put the "mechanize" folder. Write this in pymol to see the paths where pymol is searching for "mechanize"
# import sys; print(sys.path)
#-------------------------------------------------------------------------------
"""
Example for pymol script to start the functions. For example: trypropka.pml
Execute with pymol or start pymol and: File->Run->trypropka.pml
##############################################################################################################################################################################################################################
### Point to your directory with the script.
#cd /homes/linnet/Documents/Speciale/5NT-project/Mutant-construct/predict_reactivity/propka
cd C:\Users\tlinnet\Documents\My Dropbox\Speciale\5NT-project\Mutant-construct\predict_reactivity\propka
### If you have the script in your working directory the
#run ./propka.py
### You can also make the script general available. Put it into your python path. Ex: C:\Program Files (x86)\PyMOL\PyMOL\modules Then do instead:
import propka
### The fastest method is just to write propka. Then the pymol "all" molecule is assumed and send to server. But best to write molecule name
fetch 4ins, async=0
#propka
propka 4ins
### Fetch 4ins from web. async make sure, we wait to have completed
#fetch 4ins, async=0
#propka molecule=4ins, chain=*, resi=19.20, resn=ASP.TYR, logtime=_, verbose=yes, showresult=no
### If there is problems to get the mechanize module working, get the result .pka file at the propka webpage manually.
### Then run with method=file and point to .pka file.
#fetch 4ins, async=0
#hide everything, 4ins
#show cartoon, 4ins
#propka method=file, molecule=4ins, resi=18.25-30, resn=cys, file_name=.\Results_propka\4ins_.pka
### Some more examples. This molecule has 550 residues, so takes a longer time.
#fetch 1hp1, async=0
#propka molecule=1hp1, chain=A, resi=308.513, resn=CYS, logtime=_, verbose=no, showresult=no
### One can also just make a lookup for a protein. Use function: getpropka
### Note. This does only print the result to the pymol command line
#getpropka source=ID, PDBID=4ake, logtime=_, server_wait=3.0, verbose=no, showresult=yes
#getpropka source=ID, PDBID=4ins, logtime=_, server_wait=3.0, verbose=yes, showresult=no
#########input paramaters#########
# method : method=upload is default. This sends .pdb file and request result from propka server.
## method=file will only process a manual .pka file, and write a pymol command file. No use of mechanize.
# molecule : name of the molecule. Ending of file is assumed to be .pdb
# chain : which chains are saved to file, before molecule file is send to server. Separate with "." Ex: chain=A.b
# resi : Select by residue number, which residues should be printed to screen and saved to the log file: \Results_propka\_Results.log.
## Separate with "." or make ranges with "-". Ex: resi=35.40-50
# resn : Select by residue name, which residues should be printed to screen and saved to the log file: \Results_propka\_Results.log.
## Separate with "." Ex: resn=cys.tyr
# logtime : Each execution give a set of files with the job id=logtime. If logtime is not provided, the current time is used.
## Normal it usefull to set it empty. Ex: logtime=_
# verbose : Verbose is switch, to turn on messages for the mechanize section. This is handsome to see how mechanize works, and for error searching.
# showresult : Switch, to turn on all results in pymol command window. Ex: showresult=yes
# file_name: only used for: propka method=file . Write the path to .pka file
# server_wait=3.0s is default. This defines how long time between asking the server for a result. Set no lower than 3 seconds.
# version=v3.1 is default. This is what version of propka which would be used.
## Possible: 'v3.1','v3.0','v2.0'. If a newer version is available than the current v3.1, a error message is raised to make user update the script.
# source : source=upload is default and is set at the propka webpage. Can only be used for in the function "getpropka".
# getpropka source=ID, PDBID=4ake , one can print to the command line, the pka value for any official pdb ID. No files are displayed in pymol.
##############################################################################################################################################################################################################################
"""
try: from pymol import cmd; runningpymol='yes'
except: runningpymol='no'; pass
import time, platform, os
def propka(molecule="all",chain="*",resi="0",resn="NIL",method="upload",logtime=time.strftime("%m%d",time.localtime()),server_wait=3.0,version="v3.1",verbose="no",showresult="no",file_name="NIL"):
Script_Version="20110816"
### First we have to be sure, we give reasonable arguments
assert method in ['upload', 'file'], "'method' has to be either: method=upload or method=file"
assert molecule not in ['NIL'], "You always have to provide molecule name. Example: molecule=4ins"
### If molecule="all", then try to get the first molecule
if molecule=="all":
assert len(cmd.get_names())!=0, "Did you forget to load a molecule? There are no objects in pymol."
molecule=cmd.get_names()[0]
### To print out to screen for selected residues. Can be separated with "." or make ranges with "-". Example: resi="4-8.10"
if resi != "0": resi_range = ResiRange(resi)
else: resi_range=resi
### Also works for residue names. They are all converted to bigger letters. Example: resn="cys.Tyr"
if resn != "NIL": resn_range = ResnRange(resn); resn_range = [resn.upper() for resn in resn_range]
else: resn_range = resn
### Make chain range, and upper case.
chain = ChainRange(chain)
### Make result directory. We also the absolut path to the new directory.
Newdir = createdirs()
if method=="upload":
### We try to load mechanize. If this fail, one can always get the .pka file manual and the run: method=file
try: import mechanize; importedmechanize='yes'
except ImportError: print("Import error. Is a module missing?"); print(sys.exc_info()); print("Look if missing module is in your python path\n%s")%sys.path;importedmechanize='no'; import mechanize
### The name for the new molecule
newmolecule = "%s%s"%(molecule,logtime)
### Create the new molecule from original loaded and for the specified chains. Save it, and disable the old molecule.
cmd.create("%s"%newmolecule, "%s and chain %s"%(molecule,chain))
cmd.save("%s%s.pdb"%(Newdir,newmolecule), "%s"%newmolecule)
cmd.disable("%s"%molecule)
if molecule=="all": cmd.enable("%s"%molecule); cmd.show("cartoon", "%s"%molecule)
### Let the new molecule be shown in cartoon.
cmd.hide("everything", "%s"%newmolecule)
cmd.show("cartoon", "%s"%newmolecule)
### Make the absolut path to the newly created .pdb file.
PDB="%s%s.pdb"%(Newdir,newmolecule);source="upload"; PDBID=""
### Request server, and get the absolut path to the result file.
file_name = getpropka(PDB,chain,resi,resn,source,PDBID,logtime,server_wait,version,verbose,showresult)
### Open the result file and put in into a handy list.
list_results = importpropkaresult(file_name)
### Now we check if the script is actually the newest one.
try: checkversion(Script_Version,verbose)
except cmd.Warning: pass
if method=="file":
assert file_name not in ['NIL'], "You have to provide path to file. Example: file_name=./Results_propka/4ins_2011.pka"
assert ".pka" in file_name, 'The propka result file should end with ".pka" \nExample: file_name=./Results_propka/4ins_2011.pka \nfilename_name=%s'%(file_name)
### The name for the molecule we pass to the writing script of pymol commands
newmolecule = "%s"%molecule
### We open the result file we have got in the manual way and put in into a handy list.
list_results = importpropkaresult(file_name)
### Then we print the interesting residues to the screen.
printpropkaresult(list_results, resi, resi_range, resn, resn_range, showresult)
### Now create the pymol command file. This should label the protein. We get back the absolut path to the file, so we can execute it.
result_pka_pymol_name = writepymolcmd(newmolecule,file_name)
### Now run our command file. But only if we are running pymol.
if runningpymol=='yes': cmd.do("run %s"%result_pka_pymol_name)
return()
if runningpymol !='no': cmd.extend("propka",propka)
def getpropka(PDB="NIL",chain="*",resi="0",resn="NIL",source="upload",PDBID="",logtime=time.strftime("%Y%m%d%H%M%S",time.localtime()),server_wait=3.0,version="v3.1",verbose="no",showresult="no"):
try: import mechanize; importedmechanize='yes'
except ImportError: print("Import error. Is a module missing?"); print(sys.exc_info()); print("Look if missing module is in your python path \n %s"%sys.path);importedmechanize='no'
propka_v_201108 = 3.1
url = "http://propka.ki.ku.dk/"
assert version in ['v2.0', 'v3.0', 'v3.1'], "'version' has to be either: 'v2.0', 'v3.0', 'v3.1'"
assert source in ['ID', 'upload', 'addr', 'input_file'], "'source' has to be either: 'ID', 'upload', 'addr', 'input_file'"
if source=="upload": assert PDB not in ['NIL'], "You always have to provide PDB path. Example: PDB=.\Results_propka\4ins2011.pdb"
if source=="ID": assert len(PDBID)==4 , "PDBID has to be 4 characters"
### To print out to screen for selected residues. Can be separated with "." or make ranges with "-". Example: resi="4-8.10"
if resi != "0": resi_range = ResiRange(resi)
else: resi_range=resi
### Also works for residue names. They are all converted to bigger letters. Example: resn="cys.Tyr"
if resn != "NIL": resn_range = ResnRange(resn); resn_range = [resn.upper() for resn in resn_range]
else: resn_range = resn
### Start the browser
br = mechanize.Browser()
### We pass to the server, that we are not a browser, but this python script. Can be used for statistics at the propka server.
br.addheaders = [('User-agent', 'pythonMechanizeClient')]
### To turn on debugging messages
#br.set_debug_http(True)
### To open the start page.
page_start = br.open(url)
read_start = page_start.read()
if verbose == 'yes': print(br.title()); print(br.geturl())
### To get available forms
page_forms = [f.name for f in br.forms()]
if verbose == 'yes': print(page_forms)
### Select first form
br.select_form(name=page_forms[0])
## Print the current selected form, so we see that we values we start with.
if verbose == 'yes': print(br.form)
### Print the parameters of the 'version' RadioControl button and current value
if verbose == 'yes': print(br.find_control(name='version')), br.find_control(name='version').value
### This is to check, that the current script is "up-to-date".
propka_v_present = float(br.find_control(name='version').value[0].replace('v',''))
if propka_v_present > propka_v_201108:
raise UserWarning('\nNew version of propka exist.\nCheck/Update your script.\nPresent:v%s > Script:v%s'%(propka_v_present,propka_v_201108))
### Change the parameters of the 'version' radio button and then reprint the new value. Input has to be in a list [input].
br.form['version'] = [version]
if verbose == 'yes': print(br.find_control(name='version').value)
### Print the parameters of the 'source' RadioControl button and current value
if verbose == 'yes': print(br.find_control(name='source'), br.find_control(name='source').value)
### Change the parameters of the 'source' radio button and then reprint the new value. Input has to be in a list [input].
br.form['source'] = [source]
if verbose == 'yes': print(br.find_control(name='source').value)
### This step was the must strange and took a long time. For finding the information and the double negative way.
### One have to enable the pdb button. Read more here: http://wwwsearch.sourceforge.net/old/ClientForm/ ("# All Controls may be disabled.....)
PDBID_control = br.find_control("PDBID")
PDB_control = br.find_control("PDB")
if verbose == 'yes': print(PDBID_control.disabled, PDB_control.disabled)
if source == "ID": PDBID_control.disabled=False; PDB_control.disabled=True
if source == "upload": PDBID_control.disabled=True; PDB_control.disabled=False
if verbose == 'yes': print(PDBID_control.disabled, PDB_control.disabled)
### We create the result dir, and take with us the 'path' to the result dir.
Newdir = createdirs()
### Open all the files, and assign them.
if source == "upload": filename = PDB
if source == "ID": filename = PDBID
files = openfiles(Newdir, filename, logtime, source)
result_pka_file=files[0];result_input_pka_file=files[1];result_log=files[2];filepath=files[3];result_pka_file_name=files[4];result_pka_file_stripped=files[5]
## Print the parameters of the 'PDBID' TextControl button and current value
if source == "ID" and verbose == 'yes': print(br.find_control(name='PDBID')); print(br.find_control(name='PDBID').value)
## Change the parameters of the 'PDBID' TextControl and then reprint the new value. Input has just to be a string.
if source == "ID": br.form["PDBID"] = PDBID
if source == "ID" and verbose == 'yes': print(br.find_control(name='PDBID').value)
## Print the parameters of the 'PDB' TextControl button and current value
if source == "upload" and verbose == 'yes': print(br.find_control(name='PDB')); print(br.find_control(name='PDB').value)
## Change the parameters of the 'PDB' FileControl and then reprint the new value. Input has just to be a string.
if source == "upload": PDBfilename=PDB; PDBfilenamepath=PDB
if source == "upload": br.form.add_file(open(PDBfilename), 'text/plain', PDBfilenamepath, name='PDB')
if source == "upload" and verbose == 'yes': print(br.find_control(name='PDB')); print(br.find_control(name='PDB').value)
## Now reprint the current selected form, so we see that we have the right values.
if verbose == 'yes': print(br.form)
### Make "how" we would like the next request. We would like to "Click the submit button", but we have not opened the request yet.
req = br.click(type="submit", nr=0)
### Have to pass by a mechanize exception. Thats the reason for the why True
### The error was due to: br.open(req)
###### mechanize._response.httperror_seek_wrapper: HTTP Error refresh: The HTTP server returned a redirect error that would lead to an infinite loop.
###### The last 30x error message was:
###### OK
### I haven't been able to find the refresh problem or extend the time. So we make a pass on the raised exception.
try:
print("Now sending request to server")
br.open(req)
### If there is raised an exception, we jump through to the result page after some sleep.
except mechanize.HTTPError:
### We can extract the jobid from the current browser url.
jobid = br.geturl()[32:-5]
### We notice how the script at the server presents the final result page.
url_result = url + "pka/" + jobid + ".html"
### Now we continue to try to find the result page, until we have succes. If page doesn't exist, we wait a little.
while True:
print("Result still not there. Waiting %s seconds more"%server_wait)
time.sleep(float(server_wait))
### To pass the "break" after the exception, we make a hack, wait and then go to the result page, which is the jobid.
try:
page_result = br.open(url_result)
read_result = page_result.read()
### If we don't receive a error in getting the result page, we break out of the while loop.
break
### If the page doesn't exist yet. We go back in the while loop.
except mechanize.HTTPError:
### Wait another round
pass
### If we get a timeout, we also wait.
except mechanize.URLError:
### Wait another round
pass
htmlresult="The detailed result is now available at: %s"%br.geturl()
print(htmlresult)
read_result = br.response().read()
## Now save the available links from the current page. But only links that satisfy the expression.
links_result = []
for l in br.links(url_regex='http://propka.ki.ku.dk/pka'):
links_result.append(l)
### Now follow the link to the .propka_input resultpage
if len(links_result) > 1: br.follow_link(links_result[1])
### Now get the page text for the current link
if len(links_result) > 1: read_result1 = br.response().read()
### Save the result
if len(links_result) > 1: result_input_pka_file.write(read_result1)
### Now follow the link to the .pka resultpage
if len(links_result) > 1: br.back()
result_input_pka_file.close()
### Now follow first link. "Should be" available for all versions of propka.
br.follow_link(links_result[0])
### Now get the page for the current link
read_result0 = br.response().read()
### Save the result and close file.
result_pka_file.write(read_result0)
result_pka_file.close()
### Now get the result in a list, which is sorted
list_results = importpropkaresult(result_pka_file_name)
### Print to log file
result_log.write("# executed: %s \n# logtime: %s \n# source=%s \n# PDB=%s \n# chain=%s \n# PDBID=%s \n# server_wait=%s version=%s verbose=%s showresult=%s \n# resi=%s resn=%s\n# %s \n"%(time.strftime("%Y%m%d%H%M%S",time.localtime()),logtime,source,PDB,chain,PDBID,server_wait,version,verbose,showresult,resi,resn,htmlresult))
### Print to screen
printpropkaresult(list_results, resi, resi_range, resn, resn_range, showresult)
### Now write to log
for l in list_results:
if resi != "0" and int(l[1]) in resi_range:
result_log.write("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]) + '\n')
if resn != "NIL" and l[0] in resn_range and int(l[1]) not in resi_range:
result_log.write("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]) + '\n')
result_pka_file_stripped.write("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]) + '\n')
result_pka_file_stripped.close()
result_log.close()
return(result_pka_file_name)
if runningpymol !='no': cmd.extend("getpropka",getpropka)
def openpymolfiles(file_name):
result_pka_pymol_name = file_name.replace(".pka",".pml")
result_pka_pymol = open(result_pka_pymol_name, "w")
return(result_pka_pymol, result_pka_pymol_name)
def printpropkaresult(list_results, resi, resi_range, resn, resn_range, showresult):
for l in list_results:
if resi != "0" and int(l[1]) in resi_range:
if showresult != 'yes': print("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]))
if resn != "NIL" and l[0] in resn_range and int(l[1]) not in resi_range:
if showresult != 'yes': print("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]))
if showresult == 'yes': print("%3s %3s %s %6s %3s %5s %3s %4s %s"%(l[0],l[1],l[2],l[3],l[4],l[5],l[6],l[7],l[8]))
def importpropkaresult(result_pka_file_name):
result_pka_file = open(result_pka_file_name, "r")
list_results = []
for l in result_pka_file:
if not l.strip():
continue
else:
### To search for the right lines
if l.strip().split()[0] in ['ASP', 'GLU', 'ARG', 'LYS', 'HIS', 'CYS', 'TYR', 'C-', 'N+'] and len(l.strip().split())>20:
list_results.append([l.strip().split()[0], l.strip().split()[1], l.strip().split()[2], l.strip().split()[3], l.strip().split()[4], l.strip().split()[6], l.strip().split()[7], l.strip().split()[8], l.strip().split()[9]])
### Sort the result after the residue number and then chain.
list_results.sort(key=lambda residue: int(residue[1]))
list_results.sort(key=lambda chain: chain[2])
result_pka_file.close()
return(list_results)
def createdirs():
if platform.system() == 'Windows': Newdir = os.getcwd()+"\Results_propka\\"
if platform.system() == 'Linux': Newdir = os.getcwd()+"/Results_propka/"
if not os.path.exists(Newdir): os.makedirs(Newdir)
return(Newdir)
def openfiles(Newdir, filename, logtime, source):
if source == "upload":
result_pka_file_name = filename.replace(".pdb",".pka")
result_pka_input_file_name = filename.replace(".pdb",".propka_input")
result_log_name = "%s_Results.log"%(Newdir)
result_pka_file_stripped_name = filename.replace(".pdb",".stripped")
if source == "ID":
result_pka_file_name = "%s%s%s.pka"%(Newdir,filename,logtime)
result_pka_input_file_name = "%s%s%s.propka_input"%(Newdir,filename,logtime)
result_log_name = "%s_Results.log"%(Newdir)
result_pka_file_stripped_name = "%s%s%s.stripped"%(Newdir,filename,logtime)
if platform.system() == 'Windows': filepath = "\\"
if platform.system() == 'Linux': filepath = "/"
### Open the files
result_pka_file = open(result_pka_file_name, "w")
result_input_pka_file = open(result_pka_input_file_name, "w")
result_log = open(result_log_name, "a")
result_pka_file_stripped = open(result_pka_file_stripped_name, "w")
return(result_pka_file, result_input_pka_file, result_log, filepath, result_pka_file_name,result_pka_file_stripped)
def ResiRange(resi):
resi = resi.split('.')
resiList = []
for i in resi:
if '-' in i:
tmp = i.split('-')
resiList.extend(range(int(tmp[0]),int(tmp[-1])+1))
if '-' not in i:
resiList.append(int(i))
return(resiList)
def ResnRange(resn):
resn = resn.split('.')
return(resn)
def ChainRange(chain):
chainstring = chain.replace(".","+").upper()
return(chainstring)
def writepymolcmd(newmolecule,file_name):
list_results = importpropkaresult(file_name)
files_pka_pymol = openpymolfiles(file_name)
result_pka_pymol=files_pka_pymol[0];result_pka_pymol_name=files_pka_pymol[1]
dictio = {'ASP':'CG', 'GLU':'CD', 'ARG':'CZ', 'LYS':'NZ', 'HIS':'CG', 'CYS':'SG', 'TYR':'OH', 'C-':'C', 'N+':'N'}
dictio2 = {'ASP':'D', 'GLU':'E', 'ARG':'R', 'LYS':'K', 'HIS':'H', 'CYS':'C', 'TYR':'Y', 'C-':'C-', 'N+':'N+'}
### Now start write to the file.
### Try to make silent, and not auto zoom the new objects
result_pka_pymol.write("cmd.feedback('disable','all','actions')\n")
result_pka_pymol.write("cmd.feedback('disable','all','results')\n")
result_pka_pymol.write("cmd.set('label_font_id','12')\n")
result_pka_pymol.write("cmd.set('label_size','-0.5')\n")
result_pka_pymol.write("cmd.set('label_color','grey')\n")
result_pka_pymol.write("cmd.set('auto_zoom','off')\n")
### The name for the molecules are defined here
pkamolecule="pka%s"%(newmolecule)
pkalabelmolecule="%s-lab"%(pkamolecule)
### Create the group now, so they come in order.
result_pka_pymol.write("cmd.group('%s-res','%s-*')\n"%(newmolecule,newmolecule))
### Create new empty pymol pka molecules. For pka atoms and its label.
result_pka_pymol.write("cmd.create('%s','None')\n"%(pkamolecule))
result_pka_pymol.write("cmd.create('%s','None')\n"%(pkalabelmolecule))
for l in list_results:
name=dictio[l[0]];resn=dictio2[l[0]];resi=l[1];chain=l[2];pka=l[3];buried=l[4]
if "*" in pka: pka = pka.replace("*",""); comment="*Coupled residue"
else: comment=""
### Make the selection for which atom to copy
newselection = ("/%s//%s/%s/%s"%(newmolecule,chain,resi,name))
protselect = ("%s-%s%s%s"%(newmolecule,chain,resn,resi))
result_pka_pymol.write("cmd.select('%s','byres %s')\n"%(protselect,newselection))
result_pka_pymol.write("cmd.show('sticks','byres %s')\n"%(protselect))
### The temporary name
tempname = ("%s%s%s%s"%(pkamolecule,chain,resi,name))
tempnamelabel = ("%s%s%s%s"%(pkalabelmolecule,chain,resi,name))
tempselect= ("/%s//%s/%s"%(tempname,chain,resi))
tempselectlabel= ("/%s//%s/%s"%(tempnamelabel,chain,resi))
### Copy the atom, call it by the residue name
result_pka_pymol.write("cmd.create('%s','%s',quiet=1)\n"%(tempname,newselection))
### Alter the name and the b value of the newly created atom
result_pka_pymol.write("cmd.alter('%s','b=%s')\n"%(tempselect,pka))
result_pka_pymol.write("cmd.alter('%s','vdw=0.5')\n"%(tempselect))
result_pka_pymol.write("cmd.alter('%s','name=%s%s%s')\n"%(tempselect,'"',pka,'"'))
### Now create a fake label atom, and translate it
result_pka_pymol.write("cmd.create('%s','%s',quiet=1)\n"%(tempnamelabel,tempname))
movelabelxyz = (1.5,0,0)
result_pka_pymol.write("cmd.translate('[%s,%s,%s]','%s',camera=0)\n"%(movelabelxyz[0],movelabelxyz[1],movelabelxyz[2],tempnamelabel))
### Labelling alternate positions are not allowed, so we delete that attribute for the label atoms.
result_pka_pymol.write("cmd.alter('%s','alt=%s%s')\n"%(tempselectlabel,'"','"'))
result_pka_pymol.write("cmd.label('%s','text_type=%spka=%s%s%s')\n"%(tempselectlabel,'"',pka,comment,'"'))
### Now put the atoms into a bucket of atoms
result_pka_pymol.write("cmd.create('%s','%s or (%s)',quiet=1)\n"%(pkamolecule,pkamolecule,tempselect))
result_pka_pymol.write("cmd.create('%s','%s or (%s)',quiet=1)\n"%(pkalabelmolecule,pkalabelmolecule,tempselectlabel))
### Remove the temporary atoms
result_pka_pymol.write("cmd.remove('%s')\n"%(tempname))
result_pka_pymol.write("cmd.remove('%s')\n"%(tempnamelabel))
### Delete the temporary molecule/selection
result_pka_pymol.write("cmd.delete('%s')\n"%(tempname))
result_pka_pymol.write("cmd.delete('%s')\n"%(tempnamelabel))
result_pka_pymol.write("cmd.show('spheres','%s')\n"%(pkamolecule))
result_pka_pymol.write("cmd.spectrum('b','rainbow',selection='%s',minimum='0',maximum='14')\n"%(pkamolecule))
result_pka_pymol.write("cmd.alter('%s and name 99.9','vdw=0.8')\n"%(pkamolecule))
result_pka_pymol.write("cmd.show('spheres','%s and name 99.9')\n"%(pkamolecule))
result_pka_pymol.write("cmd.color('sulfur','%s and name 99.9')\n"%(pkamolecule))
result_pka_pymol.write("cmd.group('%s-res','%s-*')\n"%(newmolecule,newmolecule))
result_pka_pymol.write("cmd.set('auto_zoom','on')\n")
result_pka_pymol.write("cmd.feedback('enable','all','actions')\n")
result_pka_pymol.write("cmd.feedback('enable','all','results')\n")
result_pka_pymol.close()
return(result_pka_pymol_name)
def checkversion(Script_Version="0",verbose='no',url="http://pymolwiki.org/index.php/Propka#ScriptVersion"):
try: import mechanize; importedmechanize='yes'
except ImportError: print("Import error. Is a module missing?"); print(sys.exc_info()); print("Look if missing module is in your python path \n %s"%sys.path);importedmechanize='no'
### Start the browser
br = mechanize.Browser()
### We pass to the server, that we are not a browser, but this python script. Can be used for statistics for the server.
br.addheaders = [('User-agent', 'pythonMechanizeClient')]
### To turn on debugging messages
#br.set_debug_http(True)
### To open the start page.
page_start = br.open(url)
read_start = page_start.read()
if verbose == 'yes': print(br.title()); print(br.geturl())
read_start_lines = read_start.splitlines()
for l in read_start_lines:
if "Current_Version=" in l:
Web_Version=l.split("=")[-1]
if verbose == 'yes': print(Web_Version)
if float(Web_Version) > float(Script_Version):
raise UserWarning('\nThe author has updated the pymol propka script.\nPresent:v%s > Script:v%s \nThe new script is available at "http://pymolwiki.org/index.php/Propka" or "http://tinyurl.com/pymolpropka"'%(Web_Version,Script_Version))
return(Web_Version, Script_Version)
ScriptVersion
Current_Version=20110816
Changelog
- 20110816
- 1) Made the execution of the pymol command script silent, by only using cmd.API. This will raise a Warning, which can be ignored.
- 2) Built-in a Script version control, to inform the user if the propka script is updated on this page
- 3) The alternate attribute for the labeling atoms are reset. It was found pymol objected altering names for atoms which had alternate positions/values.
- 4) Reorganized the input order, which means that: molecule=all is default.