Map set: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
James.watney (talk | contribs) m (→Examples) |
||
| (One intermediate revision by one other user not shown) | |||
| Line 1: | Line 1: | ||
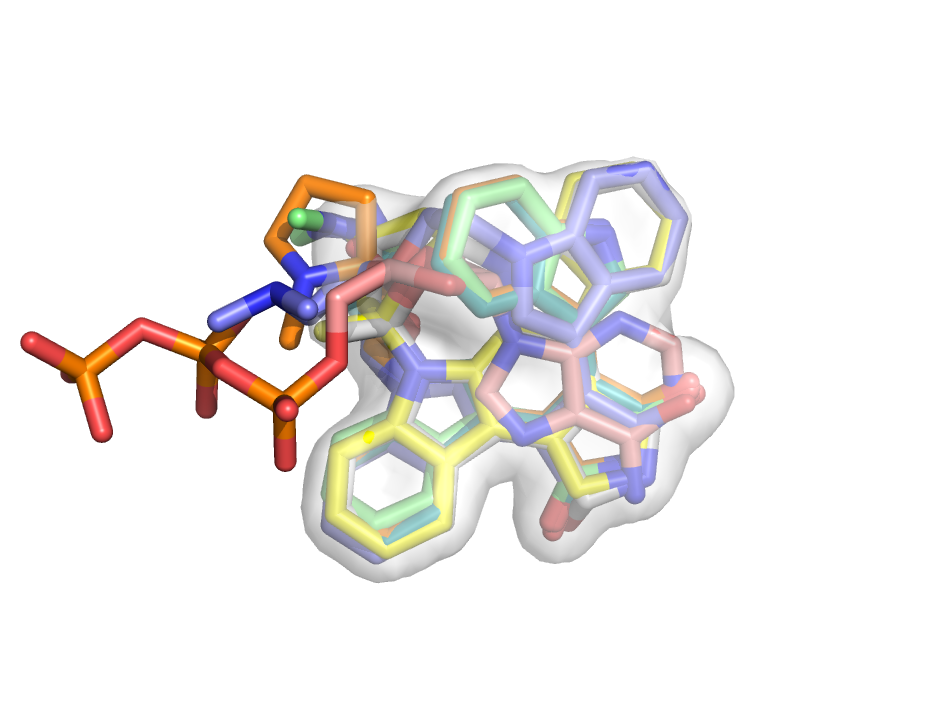
[[Image:Map_set_ex.png|right|thumb|300px|Map_set created this consensus volume representation of aligned ligands bound to kinases. See example below.]] | |||
[[Map_set]] provides a number of common operations on and between maps. For example, with [[Map_set]] you can add two maps together or create a consensus map from a series of maps or even take a difference map. | [[Map_set]] provides a number of common operations on and between maps. For example, with [[Map_set]] you can add two maps together or create a consensus map from a series of maps or even take a difference map. | ||
| Line 17: | Line 18: | ||
<source lang="python"> | <source lang="python"> | ||
# add 3 maps | # add 3 maps | ||
map_set my_sum, | map_set my_sum, sum, map1 map2 map3 | ||
# calculate the average map | # calculate the average map | ||
map_set my_avg, average, map1 map2 map3 | map_set my_avg, average, map1 map2 map3 | ||
</source> | </source> | ||
= Detailed Example = | = Detailed Example = | ||
Latest revision as of 14:38, 10 February 2011
Map_set provides a number of common operations on and between maps. For example, with Map_set you can add two maps together or create a consensus map from a series of maps or even take a difference map.
Usage
map_set name, operator, operands, target_state, source_state
operator may be,
- average
- copy
- difference
- maximum
- minimum
- sum
- unique
Examples
# add 3 maps
map_set my_sum, sum, map1 map2 map3
# calculate the average map
map_set my_avg, average, map1 map2 map3
Detailed Example
This example shows you how to create a consensus map of the bound ligand in a conserved pocket.
# fetch some similar proteins from the PDB
fetch 1oky 1h1w 1okz 1uu3 1uu7 1uu8 1uu9 1uvr, async=0
# align them all to 1oky; their ligands
# should all now be aligned
alignto 1oky
# highlight the ligands
as sticks, org
# select one of the atoms in the organic small mol
sele /1uu3//A/LY4`1374/NAT
# select entire molecules very near the chosen atom
select bm. all within 1 of (sele)
# remove the proteins; just look at small molecules
remove not (sele)
# create maps for all the ligands
python
for x in cmd.get_names():
cmd.map_new( "map_" + x, "gaussian", 0.5, x)
python end
# calculate the average map
map_set avgMap, average, map*
# show as transparent surface
set transparency, 0.5
isosurface avgSurface, avgMap, 1.0
orient vis
Notes
source_state = 0 means all states
target_state = -1 means current state
This is an experimental function.