Examples of nucleic acid cartoons: Difference between revisions
Jump to navigation
Jump to search
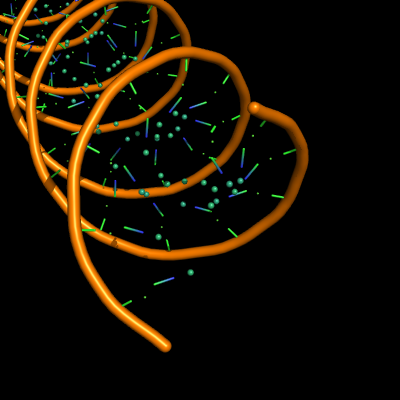
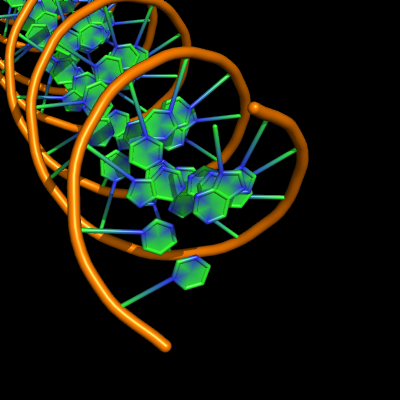
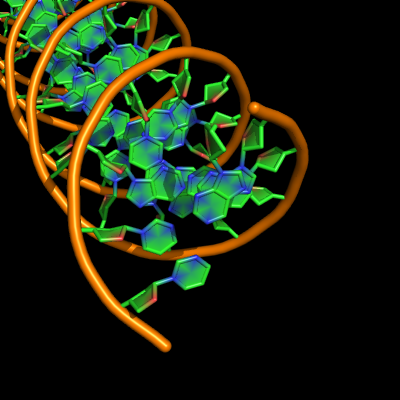
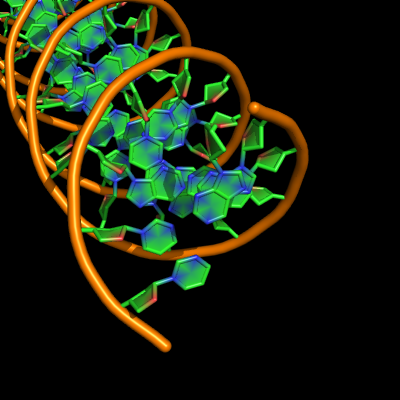
all with defaults: cartoon_ladder_mode,1 cartoon_nucleic_acid_mode,0 cartoon_ring_finder,1
| (6 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
The defaults give a phosphate backbone with single sticks passing across the full width of the base plane. | The defaults give a phosphate backbone with single sticks passing across the full width of the base plane. | ||
<source lang="python"> | <source lang="python"> | ||
set cartoon_nucleic_acid_mode, 0 # backbone follows phosphates | set cartoon_nucleic_acid_mode, 0 # backbone follows phosphates; actually Pymol itself uses setting '4' as default | ||
set cartoon_ladder_mode, 1 # sticks from backbone into nucleotide | set cartoon_ladder_mode, 1 # sticks from backbone into nucleotide | ||
set cartoon_ring_mode, 0 # no nucleotide rings | set cartoon_ring_mode, 0 # no nucleotide rings | ||
| Line 10: | Line 10: | ||
|[[Image:DNA-default-ring0-ladder1-na0-finder1.png|default view|thumb]] | |[[Image:DNA-default-ring0-ladder1-na0-finder1.png|default view|thumb]] | ||
|} | |} | ||
<!-- Prior to 4-15-09 cartoon_nucleic_acid_mode had been listed as being default value of 1 but this would go through C3, right? Fixed so value listed for cartoon_nucleic_acid_mode is | <!-- Prior to 4-15-09 cartoon_nucleic_acid_mode had been listed as being default value of 1 but this would go through C3, right? Fixed so value listed for cartoon_nucleic_acid_mode is 4 which is for following phosphates and colors the same when I look at RNA ends of 1e7k. None of the other settings looked as same as it did when opening Pymol. Any other settings don't look like the default I see when opening Pymol for RNA in 1e7k. --> | ||
== Cartoon ring mode == | == Cartoon ring mode == | ||
| Line 60: | Line 60: | ||
|align="center" | 3 || very similar to ring finder 1, slight effect on transparency = distinct behaviour? | |align="center" | 3 || very similar to ring finder 1, slight effect on transparency = distinct behaviour? | ||
|- | |- | ||
|align="center" | 4 || very similar to ring finder 1, | |align="center" | 4 || very similar to ring finder 1: finds ribose and base of nucleotides, and aromatic side chains of proteins | ||
|- | |- | ||
|align="center" | 5 || sticks visible but rings invisible | |align="center" | 5 || sticks visible but rings invisible | ||
| Line 116: | Line 116: | ||
|align="center" | 3 || appears same as mode 0? | |align="center" | 3 || appears same as mode 0? | ||
|- | |- | ||
|align="center" | 4 || appears same as mode 2? | |align="center" | 4 || appears same as mode 2? Seems to be what Pymol uses when it first opens nucleic acid containing file because any other settings change ends and colors. <!--to see this in action open 1e7k and focus on chain c--> | ||
|} | |} | ||
| Line 131: | Line 131: | ||
[[Category:Nucleic_Acids|Cartoon ring and cartoon ladder settings]] | [[Category:Nucleic_Acids|Cartoon ring and cartoon ladder settings]] | ||
[[Category:Representations]] | |||
Latest revision as of 17:31, 27 January 2010
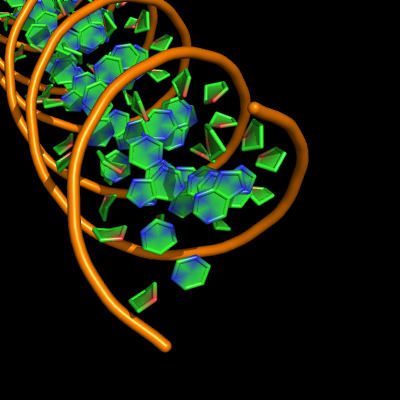
Default settings
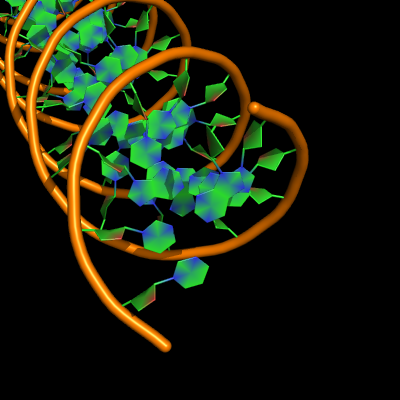
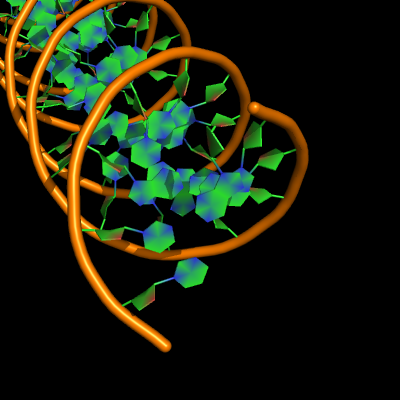
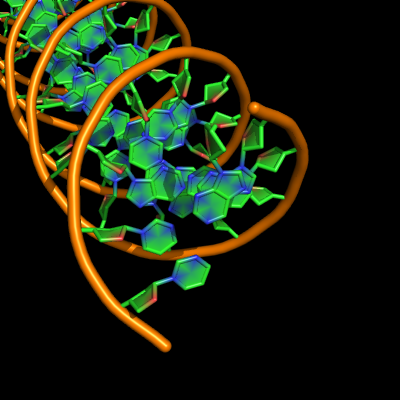
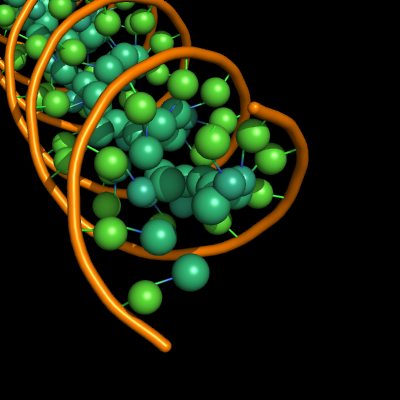
The defaults give a phosphate backbone with single sticks passing across the full width of the base plane.
set cartoon_nucleic_acid_mode, 0 # backbone follows phosphates; actually Pymol itself uses setting '4' as default
set cartoon_ladder_mode, 1 # sticks from backbone into nucleotide
set cartoon_ring_mode, 0 # no nucleotide rings
set cartoon_ring_finder, 1 # ribose and base rings (not displayed since ring mode 0)
Cartoon ring mode
Settings
set cartoon_ring_mode, value
| value | effect |
|---|---|
| 0 | stick from backbone atom to N1 of purines and N3 of pyrimidines |
| 1 | simple plane for ribose and base rings covering area between ring bonds |
| 2 | simple plane for ribose and base rings covering area inside sticks (slightly smaller than mode 1) |
| 3 | plane bounded by sticks for ribose and base rings |
| 4 | large sphere of ring diameter at centre of ribose and each base ring |
| 5 | small sphere of 1/10 diameter at centre of ribose and each base ring |
Examples
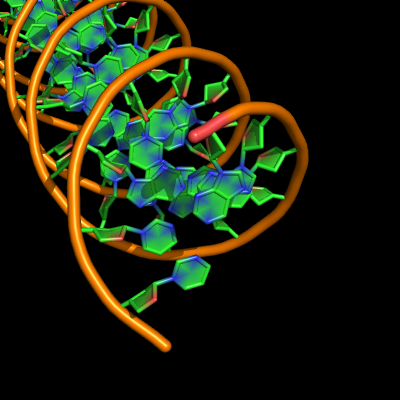
Cartoon ring finder
Settings
set cartoon_ring_finder, value
| value | effect |
|---|---|
| 0 | no rings or sticks joining them |
| 1 | both ribose and base ring |
| 2 | only base ring(s), stick connects directly from phosphate to ring |
| 3 | very similar to ring finder 1, slight effect on transparency = distinct behaviour? |
| 4 | very similar to ring finder 1: finds ribose and base of nucleotides, and aromatic side chains of proteins |
| 5 | sticks visible but rings invisible |
Examples
all with: cartoon_ladder_mode,1 cartoon_ring_mode,3 cartoon_nucleic_acid_mode,0
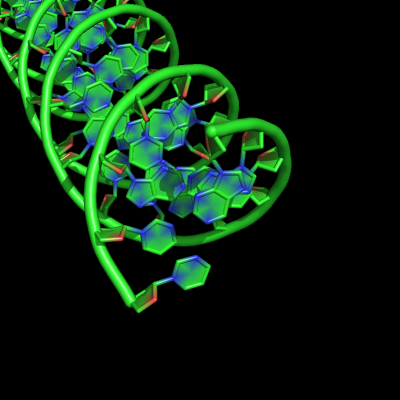
Cartoon ladder mode
Settings
set cartoon_ladder_mode, value
| value | effect |
|---|---|
| 0 | no sticks shown |
| 1 | sticks show |
Examples
all with: cartoon_ring_mode,3 cartoon_nucleic_acid_mode,0 cartoon_ring_finder,1
Note that the visibility of the ladder sticks depends on ring mode >0, ring finder >0, nucleic acid mode = 0
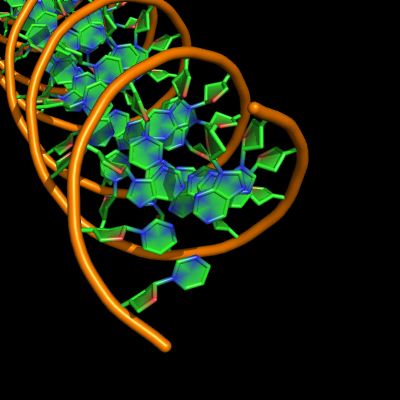
Cartoon nucleic acid mode
Settings
set cartoon_nucleic_acid_mode, value
| value | effect |
|---|---|
| 0 | smooth backbone passing through phosphorus atoms, backbone terminates at last phosphorus on either end of chain |
| 1 | smooth backbone passing through ribose C3' atoms, backbone terminates at last C3' on either end of chain |
| 2 | smooth backbone passing through phosphorus atoms, backbone terminates at last phosphorus on 5' end and O3' on 3' end (note takes O3' colour at terminus in default colouring) |
| 3 | appears same as mode 0? |
| 4 | appears same as mode 2? Seems to be what Pymol uses when it first opens nucleic acid containing file because any other settings change ends and colors. |
Examples
all with: cartoon_ladder_mode,0 cartoon_ring_mode,3 cartoon_ring_finder,1