Main Page: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
{| align="center" | |||
|+ style="font-size: | {| align="center" style="padding-bottom: 4em;" | ||
|- style="text-align:center; font-weight:bold; color: # | |+ style="font-size:210%; font-weight: bold; color:#032d45; text-align:center; padding: 5px; margin-bottom: 7px" | Welcome to the PyMOL Wiki! | ||
| | |- style="text-align:center; font-weight:bold; color: #6d6003; font-size: 140%; font-style: italic; font-family: serif;" | ||
| The community-run support site for the [http://pymol.org PyMOL] molecular viewer. | |||
|} | |} | ||
{| align="center" style=" | |||
| | {| align="center" width="45%" style="background: #EDEBD5; margin-bottom: 4em; border-bottom: 1px solid #AFB29E; border-left: 1px solid #AFB29E; border-right: 1px solid #AFB29E;" | ||
|+ style="font-size: 1.4em; font-weight: bold; color: #032d45; text-align:center; background: #5F7F96; padding-top:0.5em; padding-bottom: 0.25em; border-top: 2px solid #AFB29E; border-bottom: 1px solid #fff;" |Quick Links | |||
|- | |||
| style="font-size: 1.1em; color #61021F; padding: 0.5em 1em 0.5em 3em;"|'''[[:Category:Tutorials|Tutorials]]''' || '''[[TOPTOC|Table of Contents]]''' || '''[[:Category:Commands|Commands]]''' | |||
|- | |- | ||
| style=" | | style="font-size: 1.1em; color #61021F; padding: 0.5em 1em 0.5em 3em;"|'''[[:Category:Script_Library|Script Library]]''' || '''[[:Category:Plugins|Plugins]]''' || '''[[:Category:FAQ|FAQ]]''' | ||
|- | |- | ||
| style="font-size: 1.1em; color #61021F; padding: 0.5em 1em 0.5em 3em;"|'''[[Gallery]]''' | '''[[Covers]]''' | |||
||'''[[CheatSheet|PyMOL Cheat Sheet]]''' (''[[Media:PymolRef.pdf|PDF]]'') | |||
||'''[[GoogleSearch]]''' | |||
|} | |} | ||
{| | |||
{| width="100%" | |||
| style="vertical-align: top; width: 40%" | | |||
{| class="jtable" style="float: left; width: 90%;" | |||
|+ style="font-size: 1.4em; font-weight: bold; text-align:left; border-bottom: 2px solid #6678b1;" | News & Updates | |||
|- | |- | ||
| [[ | ! Warren | ||
| News about Warren DeLano's passing may be read on [[Warren|Warren's memorial page]]. | |||
|- | |- | ||
! PyMOLWiki | |||
| Reformatted Main Page. | |||
| | |||
|- | |- | ||
! PyMOLWiki | |||
| New — Search the PyMOLWiki via [[GoogleSearch]] | |||
| | |||
|- | |- | ||
! PyMOL | |||
| | | PyMOL is known to work under Mac OS X 10+ (Snow Leopard) using the new Fink and incentive builds. See [[MAC_Install#PyMOL_Install_from_Source.2C_Using_Fink|Installing PyMOL under Fink]]. | ||
|- | |- | ||
| | ! PyMOL | ||
| PyMOL now has a [[Set]] command for basic settings, and a [[Set_bond]] command for bond-only settings. | |||
|- | |- | ||
! PyMOL | |||
| [[Space]]— new command to control PyMOL's usage of color spaces. | |||
|} | |} | ||
| | |style="vertical-align: top; width: 40%"| | ||
{| class="jtable" style="float: right; width: 90%" | |||
|+ style="font-size: 1.4em; font-weight: bold; text-align:left; border-bottom: 2px solid #6678b1;" |Do you know this PyMOL command... | |||
| | |||
{| | |||
| | |||
|- | |- | ||
| | |<div class="didyouknow" > | ||
<dpl> | |||
namespace= | |||
category=Commands | |||
includepage=* | |||
includemaxlength=300 | |||
escapelinks=false | |||
resultsheader=__NOTOC__ __NOEDITSECTION__ | |||
randomcount=1 | |||
mode=userformat | |||
addpagecounter=true | |||
listseparators=,<h3>[[%PAGE%]]</h3>,,\n | |||
</dpl> | |||
</div> | |||
<div style="clear: both;"></div> | |||
|} | |} | ||
| | | | ||
|style="vertical-align: top; width: 18%"| | |||
<dpl> | |||
imagecontainer=Covers | |||
randomcount=1 | |||
escapelinks=false | |||
openreferences=true | |||
listseparators=[[,%PAGE%,|thumb|185px|A Random PyMOL-generated Cover. See [[Covers]].]],\n | |||
</dpl> | |||
| | |||
|- | |||
|} | |} | ||
Revision as of 13:03, 17 November 2009
| The community-run support site for the PyMOL molecular viewer. |
| Tutorials | Table of Contents | Commands |
| Script Library | Plugins | FAQ |
| Gallery | Covers | PyMOL Cheat Sheet (PDF) | GoogleSearch |
|
|
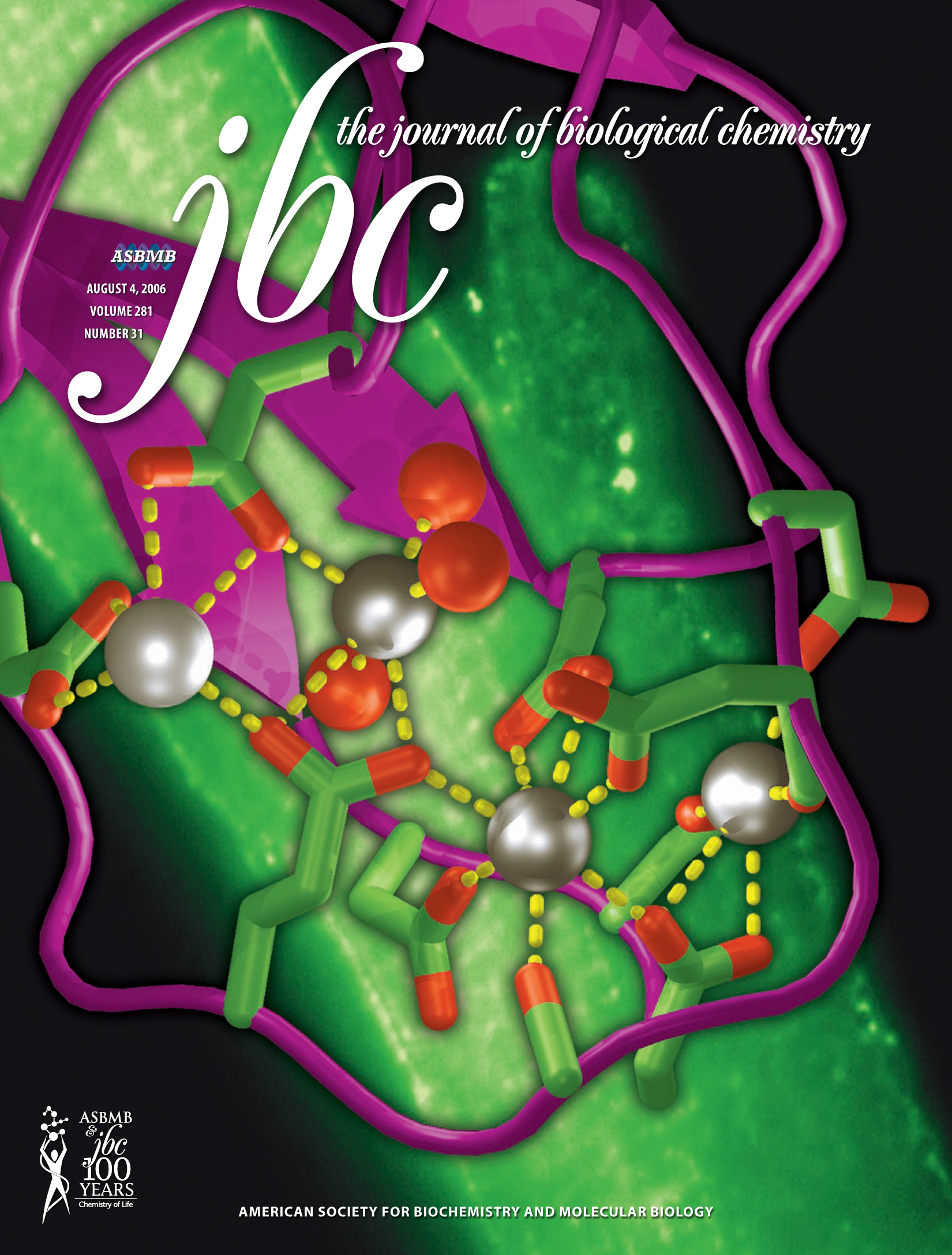 A Random PyMOL-generated Cover. See Covers. |