Nuccyl: Difference between revisions
No edit summary |
No edit summary |
||
| Line 19: | Line 19: | ||
[[Category:PyMol_Integration|Nuccyl]] | [[Category:PyMol_Integration|Nuccyl]] | ||
[[Category:Nucleic_Acids|Nuccyl]] | [[Category:Nucleic_Acids|Nuccyl]] | ||
[[Category:Biochemical_Properties]] | |||
Latest revision as of 14:15, 12 September 2008
See Nuccyl Home
Overview
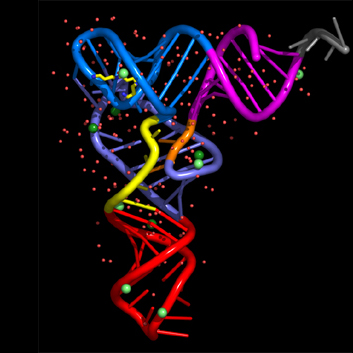
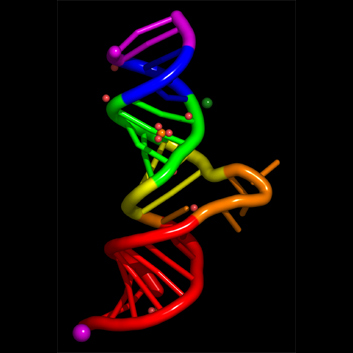
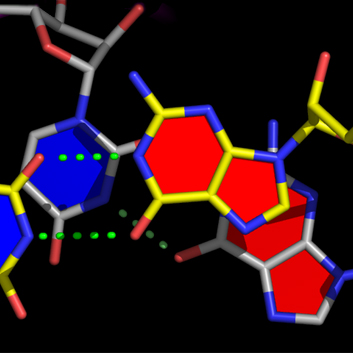
nuccyl is a Perl program that allows PyMOL, a powerful open-source molecular modeling system, to display atomic models of nucleic acids in a highly simplified representation. By depicting the nucleic acid backbone as a cartoon and the bases of nucleotides as cylinders, similarly to the excellent programs Ribbons and Drawna, nuccyl and PyMOL allow to quickly grasp the overall folding of an RNA or DNA molecule
nuccyl produces base cylinder coordinates and command files to display them with PyMOL. The base pair analysis programs RNAView and 3DNA can also be optionally used to obtain starting input files for nuccyl.
RNAView can be downloaded and installed as described; a publication describing the program is also available. Information and download links for 3DNA may be found at the program's home page; a paper has also been published.
The four example images Copyright © 2000-2005 Luca Jovine. Do NOT reuse them without contacting him or reading his release license for the images!