Protein contact potential: Difference between revisions
(New page: Protein_Contact_Potential is an automated PyMOL representation where the false red/blue charge-smoothed surface is shown on the protein. The rule of thumb with respect to PyMOL's inte...) |
No edit summary |
||
| Line 1: | Line 1: | ||
[[ | <gallery heights="200px" widths="200px" caption="Comparing PyMOL-generated electrostatics and APBS' version." align="right"> | ||
Image:Prot_contact_pot.png|PyMOL Charge-smoothed potential (see below) | |||
Image:Prot_contact_APBS.png|APBS-generated potential (see [[APBS]]). | |||
</gallery> | |||
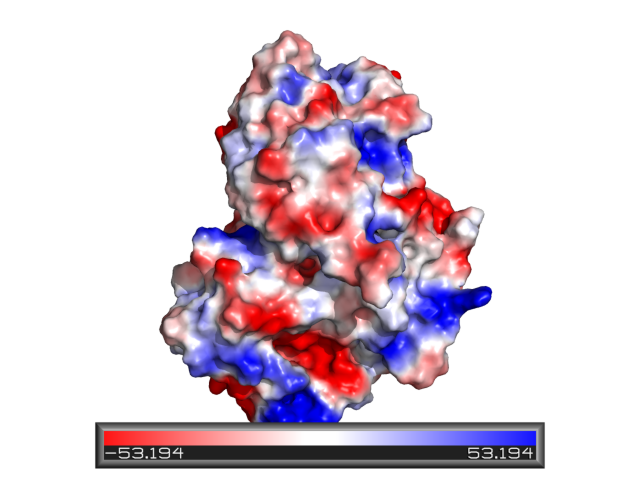
The rule of thumb with respect to PyMOL's internal "protein contact potential" is that if you care enough to be concerned with how it works, then you should instead be using a true Possion-Boltzman electrostatics | [[Protein_Contact_Potential|Protein contact potential]] is an automated PyMOL representation where the false red/blue charge-smoothed surface is shown on the protein. | ||
solver such as APBS. | |||
The rule of thumb with respect to PyMOL's internal "protein contact potential" is that if you care enough to be concerned with how it works, then you should instead be using a true Possion-Boltzman electrostatics solver such as [[APBS]]. | |||
Regardless, what PyMOL does to generate a qualitative electrostatic representation (via action '''popup->generate->vacuum electrostatics->...''') amounts to averaging charges over a small region of space using a | Regardless, what PyMOL does to generate a qualitative electrostatic representation (via action '''popup->generate->vacuum electrostatics->...''') amounts to averaging charges over a small region of space using a | ||
Revision as of 15:06, 12 September 2008
- Comparing PyMOL-generated electrostatics and APBS' version.
APBS-generated potential (see APBS).
Protein contact potential is an automated PyMOL representation where the false red/blue charge-smoothed surface is shown on the protein.
The rule of thumb with respect to PyMOL's internal "protein contact potential" is that if you care enough to be concerned with how it works, then you should instead be using a true Possion-Boltzman electrostatics solver such as APBS.
Regardless, what PyMOL does to generate a qualitative electrostatic representation (via action popup->generate->vacuum electrostatics->...) amounts to averaging charges over a small region of space using a quasi-Coulombic-shaped convolution function. Nathan Baker has suggested that we simply refer to this approach as "charge smoothing".
PyMOL's use of the term "contact" relates to the fact that the potential shown by the default coloring approximates the potential that would be felt by a point-charge one solvent radius *above* the protein surface, if we ignore solvent screening and only consider nearby atoms. My personal opinion is that this sort of treatment makes a lot sense when viewing results from Poisson-Boltmann calculations as well.
As an aside: It has never made any sense to me whatsoever that elecrostatics visualization programs would show potentials located right on the molecular surface given that (1) we employ point charge models that have *only* been parameterized to approximate potential energies computed between partial charges located at atomic centers and that (2) the molecular surface is located in the region of space where artifacts & noise in a PB calculation would be greatest due to discrete discontinuities between the low-dielectric interior and the high-dielectric exterior (solvent region).