Grid mode: Difference between revisions
Jump to navigation
Jump to search
| Line 19: | Line 19: | ||
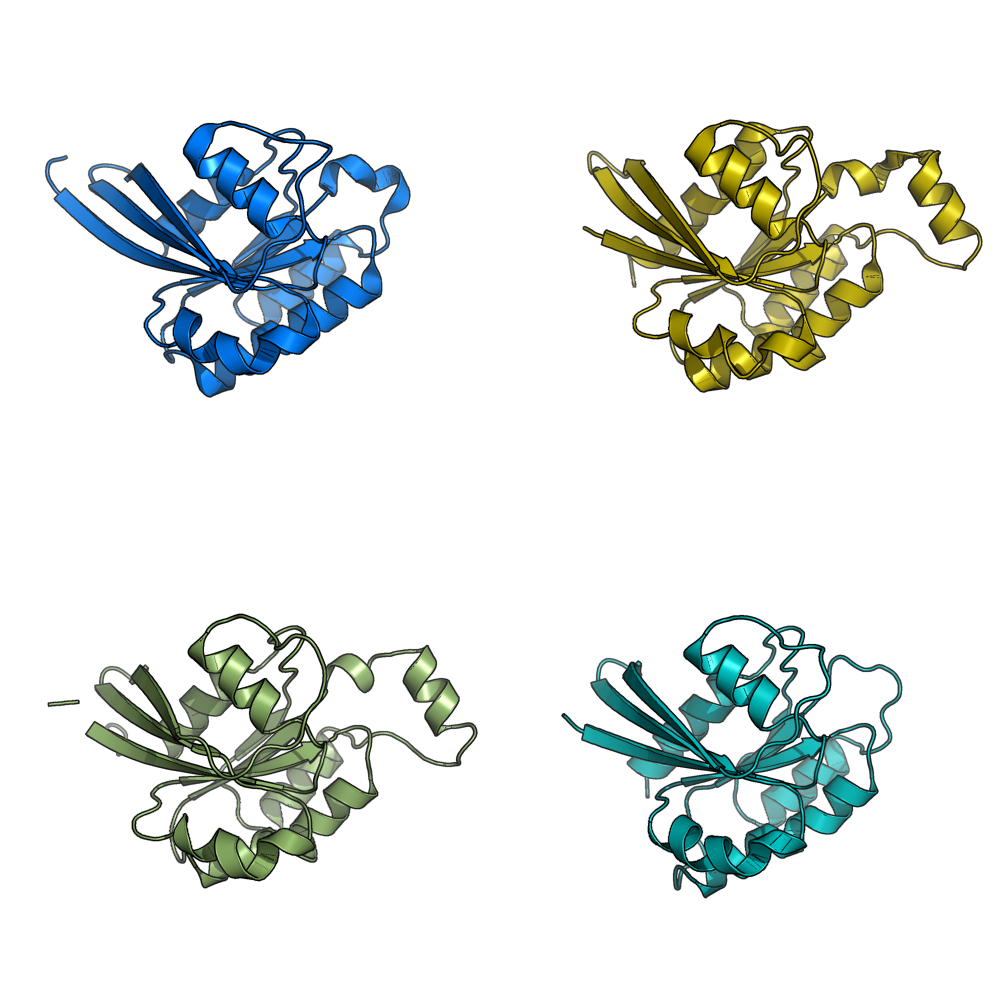
Image:Gm1.png|grid_mode on. Each molecule is in its own window. | Image:Gm1.png|grid_mode on. Each molecule is in its own window. | ||
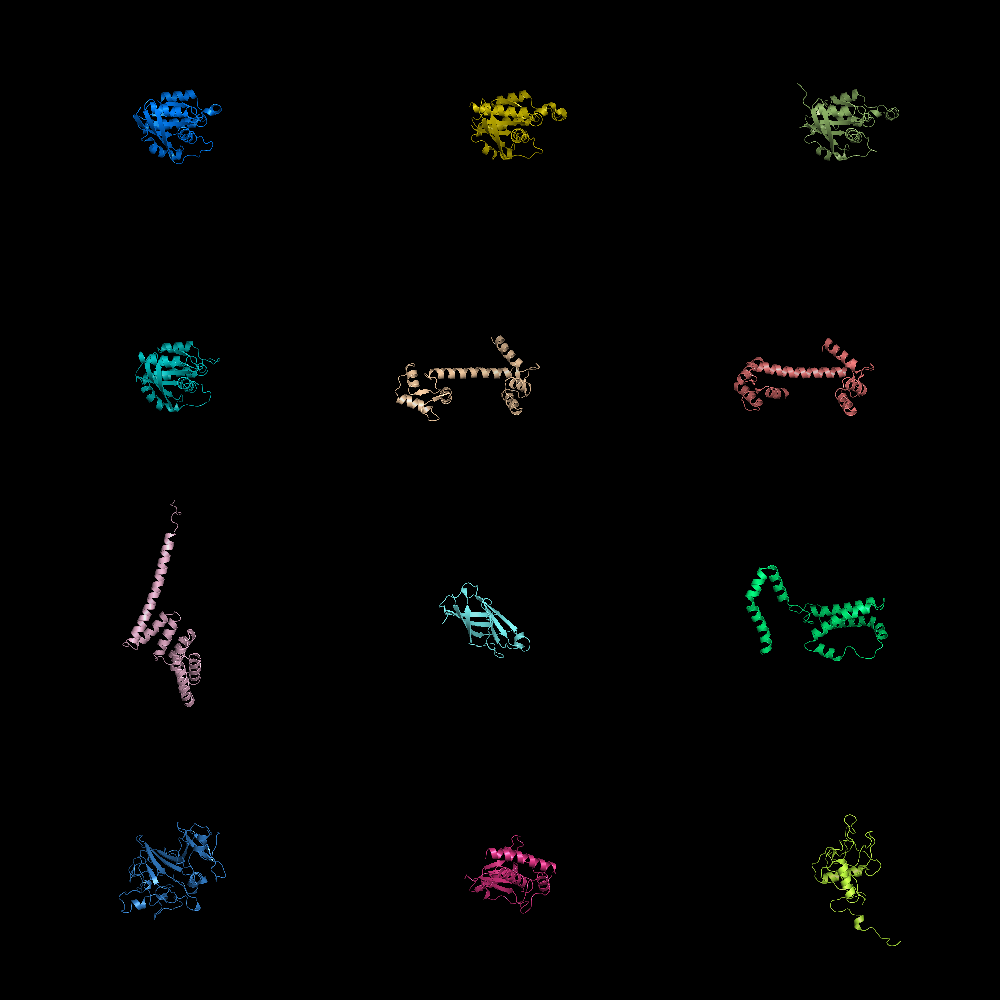
Image:Gm2.png|grid_mode on. Lots of molecules shown. | Image:Gm2.png|grid_mode on. Lots of molecules shown. | ||
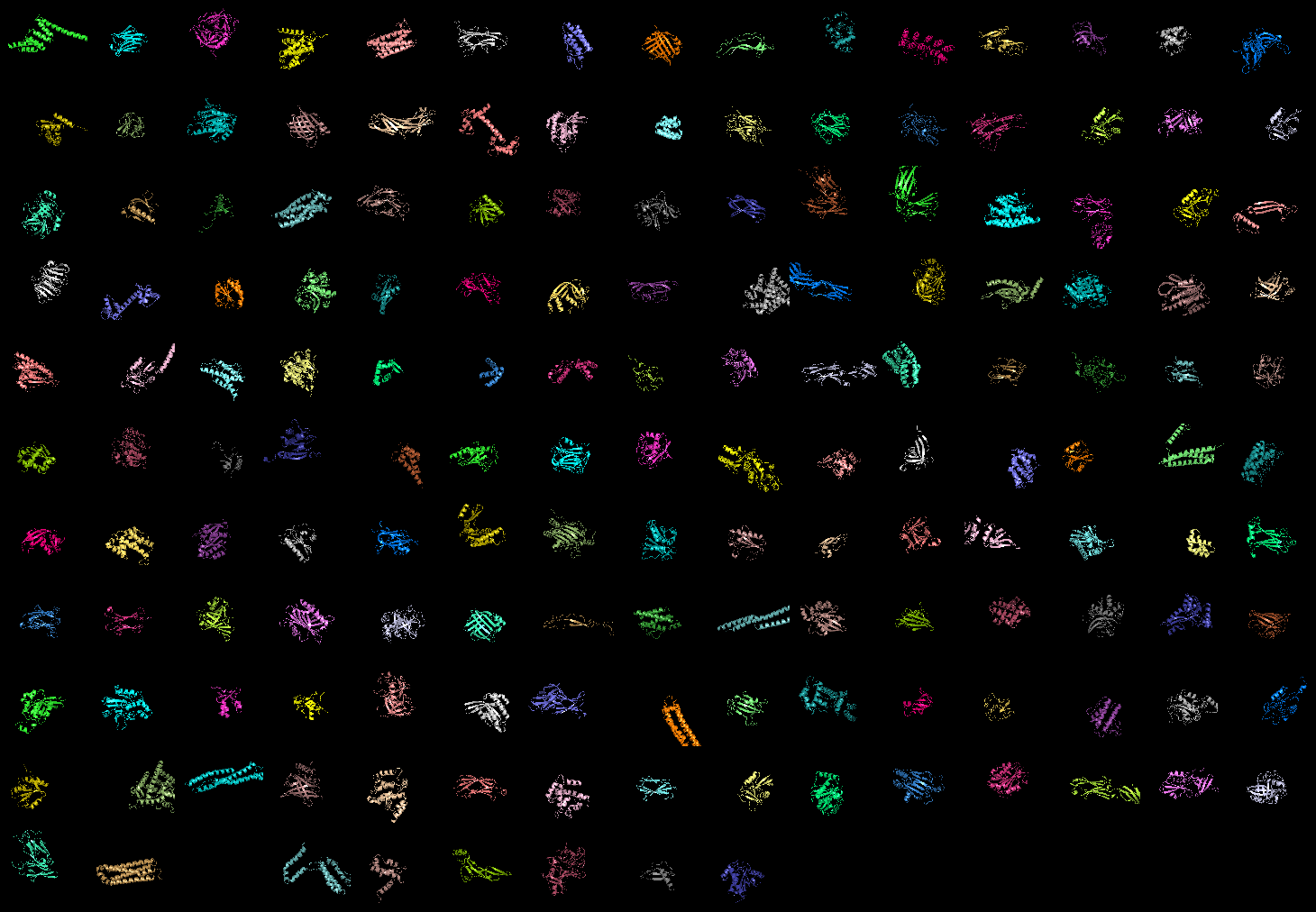
Image:Wow_grid_mode.png|grid_mode on. 159 proteins shown! | |||
</gallery> | </gallery> | ||
== See Also == | == See Also == | ||
Revision as of 13:45, 24 March 2008
Overview
grid_mode partitions the screen into a grid and displays each molecule in one grid location. Each molecule rotates, zooms, etc in that grid. A possibly very useful option for PyMOL. Each grid area is assigned a grid_slot number. This allows you to assign objects to certain grids; very helpful.
Hint: When using grid_mode with many molecules, it's sometimes good to align their centers of mass. This puts them all squarely in the middle of their grid element. The alignto command from cealign can do this for you.
Syntax
# turn on grid mode
set grid_mode,1
# turn off grid mode
set grid_mode,0