FindSurfaceCharge: Difference between revisions
Jump to navigation
Jump to search
TeddyWarner (talk | contribs) (Changed function significantly) |
TeddyWarner (talk | contribs) mNo edit summary |
||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = module | |type = module | ||
|filename = | |filename = findSurfaceCharge.py | ||
|author = [[User:TeddyWarner|Teddy Warner]] | |author = [[User:TeddyWarner|Teddy Warner]] | ||
|license = BSD-2-Clause | |license = BSD-2-Clause | ||
| Line 27: | Line 27: | ||
<source lang="python"> | <source lang="python"> | ||
run findSurfaceResiduesListCharged.py | run findSurfaceResiduesListCharged.py | ||
fetch 4FIX | fetch 4FIX | ||
findSurfaceResiduesListCharged | findSurfaceResiduesListCharged | ||
Revision as of 21:01, 14 March 2023
| Type | Python Module |
|---|---|
| Download | findSurfaceCharge.py |
| Author(s) | Teddy Warner |
| License | BSD-2-Clause |
| This code has been put under version control in the project Pymol-script-repo | |
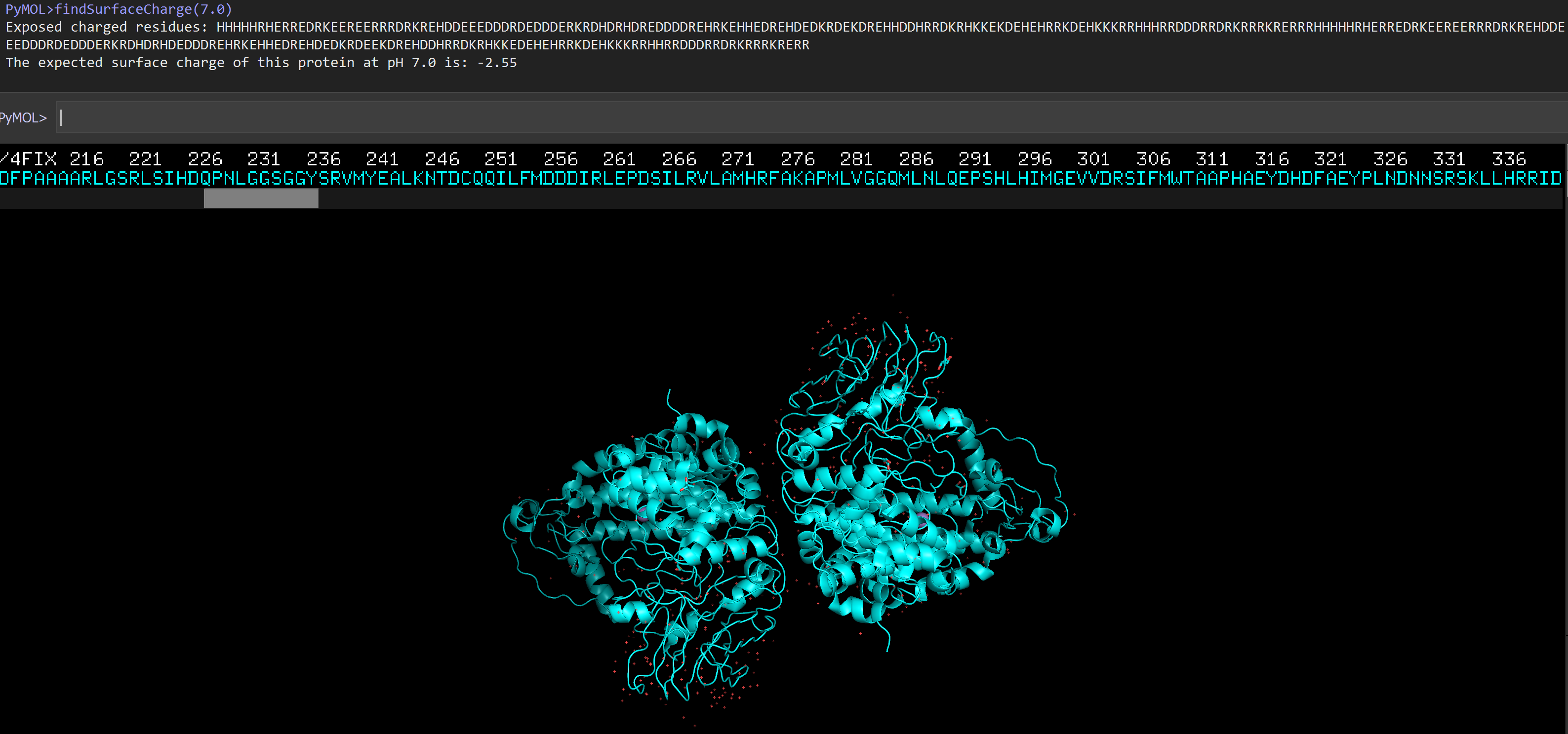
Drawing upon the findSurfaceResidues script, the findSurfaceCharge script will identify and output a list of all charged residues on the surface of a selectionand calculates the ionization state of a protein at a given pH. The charge can be calculated for either a folded or denatured protein. This function is intended to be used to give buffer conditions for mass spectrometry.
Usage
findSurfaceResiduesListCharged [pH=7.0 [, folded=True [, selection=all [, cutoff=2.5 ]]]
Arguments
- pH = float: The pH to calculate the surface charge at {default: 7.0}
- Folded = bool: Whether the program should calculate the charge of a folded (True) or denatured (False) protein.
- selection = str: The object or selection for which to find exposed residues {default: all}
- cutoff = float: The cutoff in square Angstroms that defines exposed or not. Those atoms with > cutoff Ang^2 exposed will be considered exposed {default: 2.5 Ang^2}
Examples
run findSurfaceResiduesListCharged.py
fetch 4FIX
findSurfaceResiduesListCharged
# see how pH changes the protein surface charge:
findSurfaceResiduesListCharged(7.0)
The expected surface charge of this protein at pH 7.0 is: -2.55
findSurfaceResiduesListCharged(7.0, False)
The expected charge of this denatured protein at pH 7.0 is: +0.26
findSurfaceResiduesListCharged[10.0]
The expected surface charge of this protein at pH 10.0 is: -14.19