PyMod: Difference between revisions
(Created page with " == Description == PyMod 2 is a PyMOL plugin, designed to act as simple and intuitive interface between PyMOL and several bioinformatics tools (i.e., PSI-BLAST, Clustal Omega,...") |
No edit summary |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
== Description == | == Description == | ||
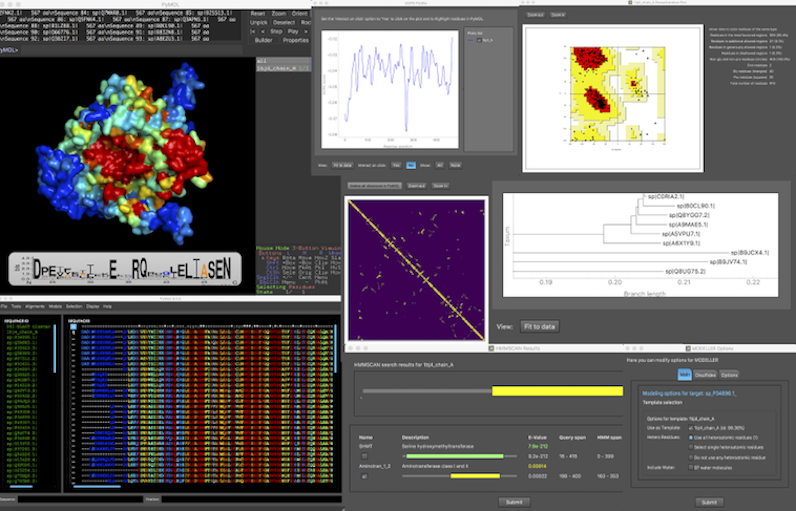
PyMod | '''PyMod 3''' is a PyMOL plugin, designed to act as simple and intuitive interface between PyMOL and several bioinformatics tools (i.e., PSI-BLAST, Clustal Omega, HMMER, MUSCLE, CAMPO, PSIPRED, and MODELLER). The current PyMod release, PyMod 3, has been extended with a rich set of functionalities that substantially improve it over its predecessor (PyMod 2.0), particularly in its ability to build homology models through the popular MODELLER package. | ||
Starting from the amino acid sequence of a target protein, users may take advantage of PyMod 3 to carry out the three steps of the homology modeling process (that is, template searching, target-template sequence alignment and model building) in order to build a 3D atomic model of a target protein (or protein complex). Additionally, PyMod 3 may also be used outside the homology modeling context, in order to extend PyMOL with numerous types of functionalities. Sequence similarity searches, multiple sequence-structure alignments and evolutionary/phylogeny analyses can all be performed in the PyMod 3/PyMOL environment. | |||
[[File:Pymod3.png|Pymod3]] | |||
== Requirements == | == Requirements == | ||
PyMod | PyMod 3 runs on Windows, macOS and Linux. It is compatible with PyMOL versions >= 2.3 and has been developed and tested on both incentive PyMOL builds (distributed by Schrodinger) and open source builds. | ||
== PyMod Download == | |||
Download: https://github.com/pymodproject/pymod/releases/download/v3.0/pymod3.zip | |||
== PyMod Install == | |||
Please refer to the PyMod 3 [https://github.com/pymodproject/pymod/releases/download/v3.0/pymod_users_guide.pdf User's Guide] to learn how to install PyMod 3 on your system. '''Long story short''': PyMod 3 can be installed as any other PyMOL plugin: | |||
1. Download the [https://github.com/pymodproject/pymod/releases/download/v3.0/pymod3.zip PyMod 3 plugin file] (a ZIP file named pymod3.zip) | |||
2. Use the PyMOL plugin manager (Plugin -> Plugin Manager from the menu of its main window) to install it | |||
3. The external tools of PyMod 3 can be obtained and configured through an easy-to-use installer dialog which can be launched by the PyMod 3 plugin (Help -> Install PyMod Components from the main menu of the PyMod plugin). The way to configure MODELLER in PyMod may vary according to your PyMOL version (in the User's Guide we cover different scenarios). The easiest way is to install Modeller using the Pymod 3 installer dialog which can be launched by the PyMod 3 plugin (a Modeller licence key is required). | |||
== How to use PyMod == | == How to use PyMod == | ||
A series of introductory tutorials and in-depth information about all the PyMod | A series of introductory tutorials and accurate, in-depth information about all the PyMod 3 functionalities can be found in its [https://github.com/pymodproject/pymod/releases/download/v3.0/pymod_users_guide.pdf User's Guide]. | ||
Documentation is also available for PyMod3 at the [https://github.com/pymodproject/pymod GitHub site]. | |||
Home page: http://schubert.bio.uniroma1.it/pymod/index.html | Home page: http://schubert.bio.uniroma1.it/pymod/index.html | ||
Download: https://github.com/pymodproject/pymod/releases/download/v3.0/pymod3.zip | |||
GitHub page: https://github.com/pymodproject/pymod | |||
== References == | |||
PyMod 3: a complete suite for structural bioinformatics in PyMOL. | |||
Janson G, Paiardini A. Bioinformatics. 2020 Oct 3:btaa849. doi: 10.1093/bioinformatics/btaa849. Online ahead of print. PMID: 33010156 | |||
PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL. | |||
Janson G, Zhang C, Prado MG, Paiardini A. Bioinformatics. 2017 Feb 1;33(3):444-446. doi: 10.1093/bioinformatics/btw638. PMID: 28158668 | |||
PyMod: sequence similarity searches, multiple sequence-structure alignments, and homology modeling within PyMOL. | |||
Bramucci E, Paiardini A, Bossa F, Pascarella S. BMC Bioinformatics. 2012 Mar 28;13 Suppl 4(Suppl 4):S2. doi: 10.1186/1471-2105-13-S4-S2. PMID: 22536966 | |||
[[Category:Plugins]] | |||
Latest revision as of 04:25, 16 November 2020
Description
PyMod 3 is a PyMOL plugin, designed to act as simple and intuitive interface between PyMOL and several bioinformatics tools (i.e., PSI-BLAST, Clustal Omega, HMMER, MUSCLE, CAMPO, PSIPRED, and MODELLER). The current PyMod release, PyMod 3, has been extended with a rich set of functionalities that substantially improve it over its predecessor (PyMod 2.0), particularly in its ability to build homology models through the popular MODELLER package.
Starting from the amino acid sequence of a target protein, users may take advantage of PyMod 3 to carry out the three steps of the homology modeling process (that is, template searching, target-template sequence alignment and model building) in order to build a 3D atomic model of a target protein (or protein complex). Additionally, PyMod 3 may also be used outside the homology modeling context, in order to extend PyMOL with numerous types of functionalities. Sequence similarity searches, multiple sequence-structure alignments and evolutionary/phylogeny analyses can all be performed in the PyMod 3/PyMOL environment.
Requirements
PyMod 3 runs on Windows, macOS and Linux. It is compatible with PyMOL versions >= 2.3 and has been developed and tested on both incentive PyMOL builds (distributed by Schrodinger) and open source builds.
PyMod Download
Download: https://github.com/pymodproject/pymod/releases/download/v3.0/pymod3.zip
PyMod Install
Please refer to the PyMod 3 User's Guide to learn how to install PyMod 3 on your system. Long story short: PyMod 3 can be installed as any other PyMOL plugin:
1. Download the PyMod 3 plugin file (a ZIP file named pymod3.zip)
2. Use the PyMOL plugin manager (Plugin -> Plugin Manager from the menu of its main window) to install it
3. The external tools of PyMod 3 can be obtained and configured through an easy-to-use installer dialog which can be launched by the PyMod 3 plugin (Help -> Install PyMod Components from the main menu of the PyMod plugin). The way to configure MODELLER in PyMod may vary according to your PyMOL version (in the User's Guide we cover different scenarios). The easiest way is to install Modeller using the Pymod 3 installer dialog which can be launched by the PyMod 3 plugin (a Modeller licence key is required).
How to use PyMod
A series of introductory tutorials and accurate, in-depth information about all the PyMod 3 functionalities can be found in its User's Guide. Documentation is also available for PyMod3 at the GitHub site.
Home page: http://schubert.bio.uniroma1.it/pymod/index.html
Download: https://github.com/pymodproject/pymod/releases/download/v3.0/pymod3.zip
GitHub page: https://github.com/pymodproject/pymod
References
PyMod 3: a complete suite for structural bioinformatics in PyMOL. Janson G, Paiardini A. Bioinformatics. 2020 Oct 3:btaa849. doi: 10.1093/bioinformatics/btaa849. Online ahead of print. PMID: 33010156
PyMod 2.0: improvements in protein sequence-structure analysis and homology modeling within PyMOL.
Janson G, Zhang C, Prado MG, Paiardini A. Bioinformatics. 2017 Feb 1;33(3):444-446. doi: 10.1093/bioinformatics/btw638. PMID: 28158668
PyMod: sequence similarity searches, multiple sequence-structure alignments, and homology modeling within PyMOL. Bramucci E, Paiardini A, Bossa F, Pascarella S. BMC Bioinformatics. 2012 Mar 28;13 Suppl 4(Suppl 4):S2. doi: 10.1186/1471-2105-13-S4-S2. PMID: 22536966