ProBiS H2O: Difference between revisions
MarkoJukic (talk | contribs) (init commit) |
m (fix download link) |
||
| (4 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = plugin | |type = plugin | ||
| | |download = http://insilab.org/probis-h2o/ | ||
|author = Marko Jukic | |author = Marko Jukic | ||
|license = | |license = | ||
| Line 8: | Line 8: | ||
== Update & Download == | == Update & Download == | ||
The most recent version can be obtained at the [http://insilab.org/ | The most recent version can be obtained at the [http://insilab.org/probis-h2o/ Insilab web page]. | ||
== Description == | == Description == | ||
| Line 23: | Line 23: | ||
or macromolecule-small molecule interfaces. For further info please refer to the relevant articles by the authors of ProBiS | or macromolecule-small molecule interfaces. For further info please refer to the relevant articles by the authors of ProBiS | ||
H2O. | H2O. | ||
| Line 95: | Line 97: | ||
that was configured using SETUP DB button): | that was configured using SETUP DB button): | ||
[[File: Define_system_tab.PNG|390 px]] | |||
because this step is optional if the user already analysed a particular protein cluster and has the | # User inputs a desired PDB ID as examined system | ||
structures already downloaded. If it is a first time experiment, the “Download” button downloads the | # User presses “Find” button to display identified structures in cluster | ||
set from the PDB website) | # User downloads all relevant structures by pressing “Download” button (this step is in parentheses because this step is optional if the user already analysed a particular protein cluster and has the structures already downloaded. If it is a first time experiment, the “Download” button downloads the set from the PDB website) | ||
# User identifies binding sites or chains of the examined system (1.) | |||
# User selects desired binding site or chain from a list | |||
# User proceeds with conserved water identification by pressing GO button | |||
| Line 109: | Line 111: | ||
After the calculation is finished ProBiS H2O switches to the cluster analysis tab and displays the results | After the calculation is finished ProBiS H2O switches to the cluster analysis tab and displays the results | ||
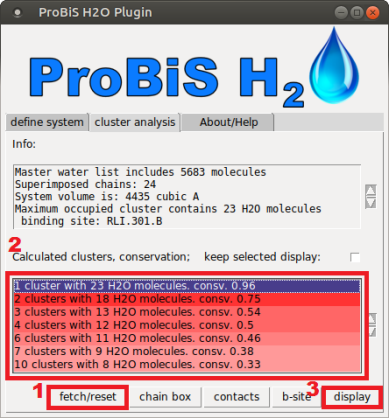
[[File: Cluster_analysis_tab.PNG|390 px]] | |||
# User displays the studied system in PyMOL display window by clicking “fetch/reset” button. This button can be used for resetting the display for alternative clustering representation | |||
# User selects the cluster from calculated clusters window | |||
# User displays the relevant cluster with display button | |||
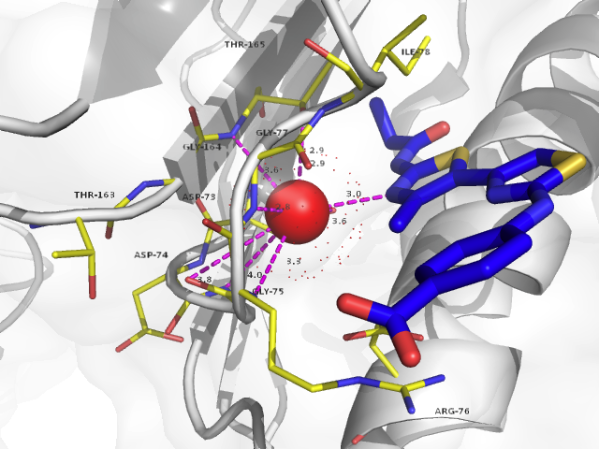
[[File: Feature 4duh.PNG|500 px]] | |||
==== About/Help Tab ==== | ==== About/Help Tab ==== | ||
Latest revision as of 09:16, 12 May 2018
| Type | PyMOL Plugin |
|---|---|
| Download | http://insilab.org/probis-h2o/ |
| Author(s) | Marko Jukic |
| License | |
Update & Download
The most recent version can be obtained at the Insilab web page.
Description
In order to help in identification of conserved water sites, we developed ProBiS H2O workflow that supports the complete process of data collection, extraction, identification of conserved water locations and result visualisation. ProBiS H2O workflow adopts the available experimental pdb data deposited in online databases or local user experimental data collections, collects similar macromolecular systems, performs local/specific binding site superimposition with the computational speed and accuracy of ProBiS algorithm, collects the experimental water location data reported in the parent macromolecule systems and transposes gathered data to the examined system, its specific chain, binding site or individual water. Water location data is then clustered to identify discrete spaces with high conservation of water molecules and visualised in the context of studied system. ProBiS H2O workflow is a robust, transparent and fast methodology with emphasis on experimental data that can identify trends in water localization intra/inter macromolecule or macromolecule-small molecule interfaces. For further info please refer to the relevant articles by the authors of ProBiS H2O.
ProBiS H2O Plugin is free for Academic (NON-COMMERCIAL) use.
For COMMERCIAL use the potential users have to write to Marko Jukic.
ProBiS H2O Plugin Features
- Graphical User Interface
- lean and fast workflow
- fast iteration
- data logging and export
- usage of custom datasets
Requirements & Dependencies
Python 2.7, NumPy (>= 1.6.1), SciPy (>= 0.9), scikit-learn (>= 1.18), PyMOL software, ProBiS (installed during ProBiS H2O setup).
The plugin works in Unix like systems. Testing environment was Ubuntu 16.04.
Installation
ProBiS H2O is installed as a PyMOL plugin:
- Run PyMOL
- Install the plugin in PyMOL by following the path: Plugin > Plugin Manager > Install New Plugin
- Restart PyMOL!
After plugin is installed, it can be run from PyMOL by following Plugin>ProBiS H2O and used through PyMOL Tcl-Tk
GUI where plugin window supplements PyMOL default control and display windows.
Important note: Before first time usage use SETUP DB button to set up th working environment.
First Time Usage
In the lower left corner of the ProBiS H2O main window resides SETUP DB button. Before usage ProBiS H2O plugin requires up-to-date information on RCSB sequence clustering and ProBiS algorithm binary file. When SETUP DB button is pressed ProBiS H2O plugin creates /Probis_H2O/ folder in default working directory of PyMOL (default is user home directory) and downloads recent sequence clustering data from RCSB PDB Database. ProBiS H2O plugin also creates /Probis_H2O/.pro/ folder where ProBiS algorithm binary file is downloaded to achieve a final workspace:
~/Probis_H2O/.pro/probis ~/Probis_H2O/clusters50.txt ~/Probis_H2O/clusters70.txt ~/Probis_H2O/clusters90.txt ~/Probis_H2O/clusters95.txt ~/Probis_H2O/bc-30.out ~/Probis_H2O/bc-40.out ~/Probis_H2O/bc-50.out ~/Probis_H2O/bc-70.out ~/Probis_H2O/bc-90.out ~/Probis_H2O/bc-95.out ~/Probis_H2O/bc-100.out
Final step is to grant probis binary executable rights:
$sudo chmod +x ~/Probis_H2O/.pro/probis
Usage
Plugin is composed of three respective tabs:
Define System Tab
Default workflow proceeds in following steps (all the data manipulation is performed in /Probis_H2O/ working directory that was configured using SETUP DB button):
- User inputs a desired PDB ID as examined system
- User presses “Find” button to display identified structures in cluster
- User downloads all relevant structures by pressing “Download” button (this step is in parentheses because this step is optional if the user already analysed a particular protein cluster and has the structures already downloaded. If it is a first time experiment, the “Download” button downloads the set from the PDB website)
- User identifies binding sites or chains of the examined system (1.)
- User selects desired binding site or chain from a list
- User proceeds with conserved water identification by pressing GO button
Cluster Analysis Tab
After the calculation is finished ProBiS H2O switches to the cluster analysis tab and displays the results
- User displays the studied system in PyMOL display window by clicking “fetch/reset” button. This button can be used for resetting the display for alternative clustering representation
- User selects the cluster from calculated clusters window
- User displays the relevant cluster with display button
About/Help Tab
The users can get reference information on the About/Help tab.
Reference
If you are using ProBiS H2O in your work, please cite:
Jukič, Marko, Janez Konc, Stanislav Gobec, Dušanka Janežič. Identification of Conserved Water Sites in Protein Structures for Drug Design. J. Chem. Inf. Model., 2017, doi: 10.1021/acs.jcim.7b00443 [1]
Licensing
ProBiS H2O software is copyrighted by :
- Marko Jukic
- Fakulteta za farmacijo
- Univerza v Ljubljani
- Askerceva 7
- SI-1000 Ljubljana
- Slovenia.
The terms stated in the following agreement apply to all files associated with the software unless explicitly disclaimed in individual files.
# Copyright Notice # ================ # # The PyMOL Plugin source code in this file is copyrighted, but you can # freely use and copy it as long as you don't change or remove any of # the copyright notices. # # ---------------------------------------------------------------------- # This PyMOL Plugin is Copyright (C) 2017 by Marko Jukic <marko.jukic@ffa.uni-lj.si> # # All Rights Reserved # # Permission to use, copy and distribute # versions of this software and its documentation for any purpose and # without fee is hereby granted, provided that the above copyright # notice appear in all copies and that both the copyright notice and # this permission notice appear in supporting documentation, and that # the name(s) of the author(s) not be used in advertising or publicity # pertaining to distribution of the software without specific, written # prior permission. # # THE AUTHOR(S) DISCLAIM ALL WARRANTIES WITH REGARD TO THIS SOFTWARE, # INCLUDING ALL IMPLIED WARRANTIES OF MERCHANTABILITY AND FITNESS. IN # NO EVENT SHALL THE AUTHOR(S) BE LIABLE FOR ANY SPECIAL, INDIRECT OR # CONSEQUENTIAL DAMAGES OR ANY DAMAGES WHATSOEVER RESULTING FROM LOSS OF # USE, DATA OR PROFITS, WHETHER IN AN ACTION OF CONTRACT, NEGLIGENCE OR # OTHER TORTIOUS ACTION, ARISING OUT OF OR IN CONNECTION WITH THE USE OR # PERFORMANCE OF THIS SOFTWARE. # ----------------------------------------------------------------------