InterfaceResidues: Difference between revisions
(Created page with '= Overview = This script finds interface residues between two proteins or chains. = Usage = <source lang="python"> interfaceResidues complexName[, chainX[, chainY ]] </source> w...') |
(select None -> 'none') |
||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
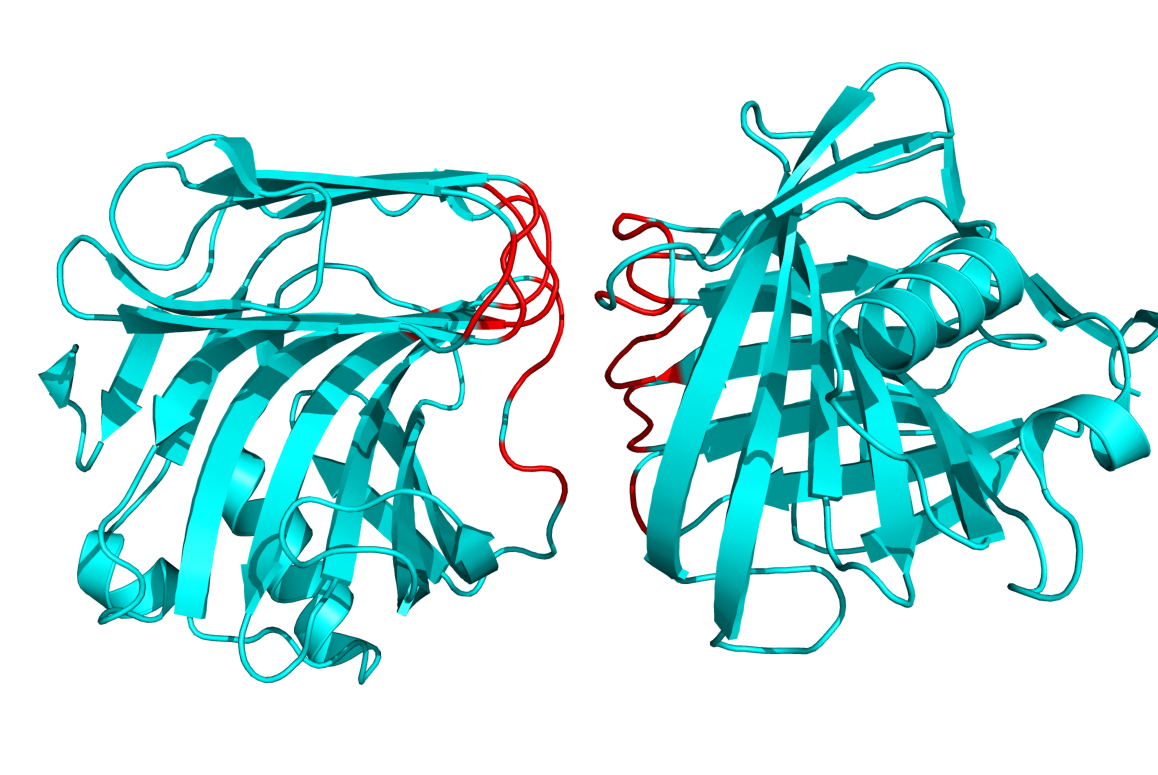
[[Image:1h4gi.png|thumb|right|300px|The red residues were detected as being in the interface of the two proteins. This is PDB 1H4G from the Example below.]] | |||
= Overview = | = Overview = | ||
This script finds interface residues between two proteins or chains. | This script finds interface residues between two proteins or chains, using the following concept. First, we take the area of the complex. Then, we split the complex into two pieces, one for each chain. Next, we calculate the chain-only surface area. Lastly, we take the difference between the comeplex-based areas and the chain-only-based areas. If that value is greater than your supplied cutoff, then we call it an interface residue. | ||
= Usage = | = Usage = | ||
<source lang="python"> | <source lang="python"> | ||
interfaceResidue complexName[, cA=firstChainName[, cB=secondChainName[, cutoff=dAsaCutoff[, selName=selectionNameToReturn ]]]] | |||
</source> | </source> | ||
where, | where, | ||
'''complexName''' | '''complexName''' | ||
:: | :: The name of the complex. cA and cB must be within in this complex | ||
''' | ''' cA''' | ||
:: | :: The name of the first chain to investigate | ||
:: | ''' cB ''' | ||
''' | :: The name of the 2nd chain to investigate | ||
:: | ''' cutoff ''' | ||
: | :: The dASA cutoff, in sqaure Angstroms | ||
''' selName ''' | |||
:: Name of the selection to return. | |||
For each residue in the newly defined interface, this script returns the model (cA or cB) name, the residue number and the change in area. To get that information use [[interfaceResidues]] in the api form as: | |||
<source lang="python"> | |||
myInterfaceResidues = interfaceResidue(complexName[, cA=firstChainName[, cB=secondChainName[, cutoff=dAsaCutoff[, selName=selectionNameToReturn ]]]]) | |||
</source> | |||
and the result, <pre>myInterfaceResidues</pre> will have all the value for you. | |||
= Examples = | |||
For these two examples, make sure you've run the script first. | |||
== Simple Example == | |||
This just finds the residues b/t chain A and chain B | |||
<source lang="python"> | |||
fetch 1h4g, async=0 | |||
interfaceResidues 1h4g | |||
</source> | |||
== More Complex Example == | |||
<source lang="python"> | |||
fetch 1qox, async=0 | |||
foundResidues = interfaceResidues("1qox", cA="c. I", cB="c. J", cutoff=0.75, selName="foundIn1QOX") | |||
</source> | |||
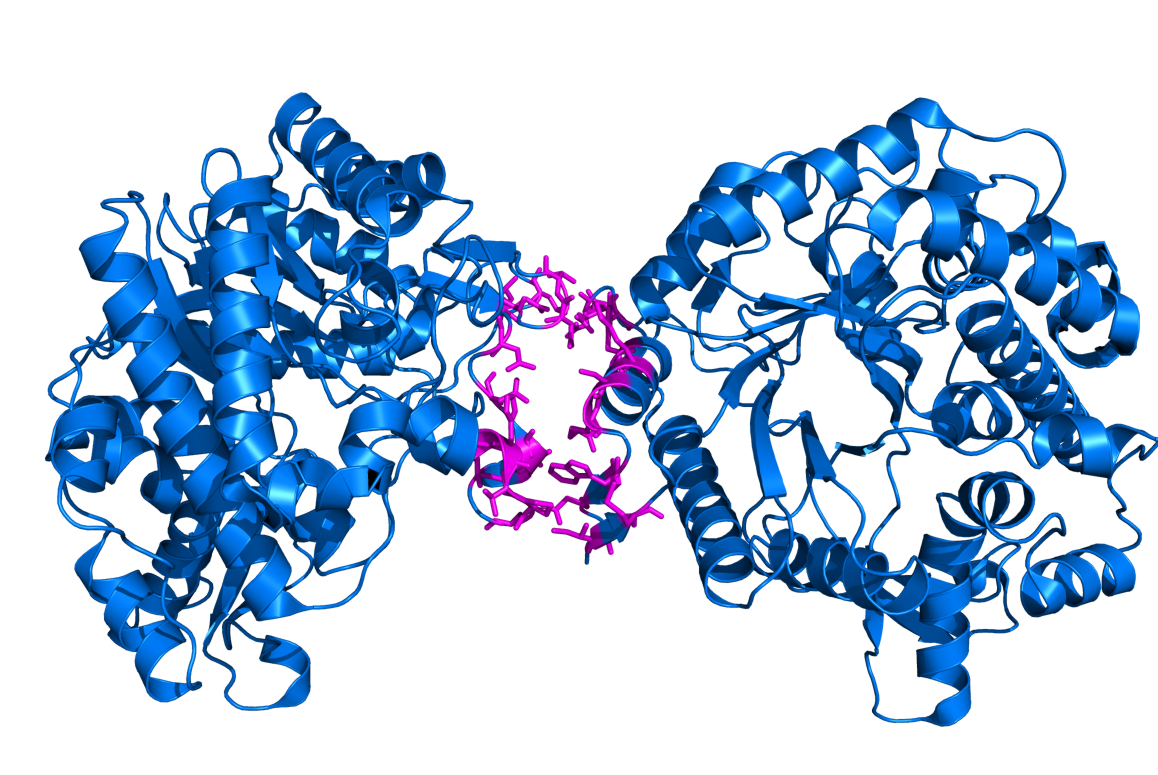
[[Image:1qoxi.png|400px|thumb|center|Interface residues between chains I and J in 1QOX.]] | |||
= The Code = | = The Code = | ||
...on | <source lang="python"> | ||
from pymol import cmd, stored | |||
def interfaceResidues(cmpx, cA='c. A', cB='c. B', cutoff=1.0, selName="interface"): | |||
""" | |||
interfaceResidues -- finds 'interface' residues between two chains in a complex. | |||
PARAMS | |||
cmpx | |||
The complex containing cA and cB | |||
cA | |||
The first chain in which we search for residues at an interface | |||
with cB | |||
cB | |||
The second chain in which we search for residues at an interface | |||
with cA | |||
cutoff | |||
The difference in area OVER which residues are considered | |||
interface residues. Residues whose dASA from the complex to | |||
a single chain is greater than this cutoff are kept. Zero | |||
keeps all residues. | |||
selName | |||
The name of the selection to return. | |||
RETURNS | |||
* A selection of interface residues is created and named | |||
depending on what you passed into selName | |||
* An array of values is returned where each value is: | |||
( modelName, residueNumber, dASA ) | |||
NOTES | |||
If you have two chains that are not from the same PDB that you want | |||
to complex together, use the create command like: | |||
create myComplex, pdb1WithChainA or pdb2withChainX | |||
then pass myComplex to this script like: | |||
interfaceResidues myComlpex, c. A, c. X | |||
This script calculates the area of the complex as a whole. Then, | |||
it separates the two chains that you pass in through the arguments | |||
cA and cB, alone. Once it has this, it calculates the difference | |||
and any residues ABOVE the cutoff are called interface residues. | |||
AUTHOR: | |||
Jason Vertrees, 2009. | |||
""" | |||
# Save user's settings, before setting dot_solvent | |||
oldDS = cmd.get("dot_solvent") | |||
cmd.set("dot_solvent", 1) | |||
# set some string names for temporary objects/selections | |||
tempC, selName1 = "tempComplex", selName+"1" | |||
chA, chB = "chA", "chB" | |||
# operate on a new object & turn off the original | |||
cmd.create(tempC, cmpx) | |||
cmd.disable(cmpx) | |||
# remove cruft and inrrelevant chains | |||
cmd.remove(tempC + " and not (polymer and (%s or %s))" % (cA, cB)) | |||
# get the area of the complete complex | |||
cmd.get_area(tempC, load_b=1) | |||
# copy the areas from the loaded b to the q, field. | |||
cmd.alter(tempC, 'q=b') | |||
# extract the two chains and calc. the new area | |||
# note: the q fields are copied to the new objects | |||
# chA and chB | |||
cmd.extract(chA, tempC + " and (" + cA + ")") | |||
cmd.extract(chB, tempC + " and (" + cB + ")") | |||
cmd.get_area(chA, load_b=1) | |||
cmd.get_area(chB, load_b=1) | |||
# update the chain-only objects w/the difference | |||
cmd.alter( "%s or %s" % (chA,chB), "b=b-q" ) | |||
# The calculations are done. Now, all we need to | |||
# do is to determine which residues are over the cutoff | |||
# and save them. | |||
stored.r, rVal, seen = [], [], [] | |||
cmd.iterate('%s or %s' % (chA, chB), 'stored.r.append((model,resi,b))') | |||
cmd.enable(cmpx) | |||
cmd.select(selName1, 'none') | |||
for (model,resi,diff) in stored.r: | |||
key=resi+"-"+model | |||
if abs(diff)>=float(cutoff): | |||
if key in seen: continue | |||
else: seen.append(key) | |||
rVal.append( (model,resi,diff) ) | |||
# expand the selection here; I chose to iterate over stored.r instead of | |||
# creating one large selection b/c if there are too many residues PyMOL | |||
# might crash on a very large selection. This is pretty much guaranteed | |||
# not to kill PyMOL; but, it might take a little longer to run. | |||
cmd.select( selName1, selName1 + " or (%s and i. %s)" % (model,resi)) | |||
# this is how you transfer a selection to another object. | |||
cmd.select(selName, cmpx + " in " + selName1) | |||
# clean up after ourselves | |||
cmd.delete(selName1) | |||
cmd.delete(chA) | |||
cmd.delete(chB) | |||
cmd.delete(tempC) | |||
# show the selection | |||
cmd.enable(selName) | |||
# reset users settings | |||
cmd.set("dot_solvent", oldDS) | |||
return rVal | |||
cmd.extend("interfaceResidues", interfaceResidues) | |||
</source> | |||
[[Category:Script_Library]] | [[Category:Script_Library]] | ||
[[Category:Structural_Biology_Scripts]] | [[Category:Structural_Biology_Scripts]] | ||
Latest revision as of 09:59, 13 November 2015
Overview
This script finds interface residues between two proteins or chains, using the following concept. First, we take the area of the complex. Then, we split the complex into two pieces, one for each chain. Next, we calculate the chain-only surface area. Lastly, we take the difference between the comeplex-based areas and the chain-only-based areas. If that value is greater than your supplied cutoff, then we call it an interface residue.
Usage
interfaceResidue complexName[, cA=firstChainName[, cB=secondChainName[, cutoff=dAsaCutoff[, selName=selectionNameToReturn ]]]]
where,
complexName
- The name of the complex. cA and cB must be within in this complex
cA
- The name of the first chain to investigate
cB
- The name of the 2nd chain to investigate
cutoff
- The dASA cutoff, in sqaure Angstroms
selName
- Name of the selection to return.
For each residue in the newly defined interface, this script returns the model (cA or cB) name, the residue number and the change in area. To get that information use interfaceResidues in the api form as:
myInterfaceResidues = interfaceResidue(complexName[, cA=firstChainName[, cB=secondChainName[, cutoff=dAsaCutoff[, selName=selectionNameToReturn ]]]])
and the result,
myInterfaceResidues
will have all the value for you.
Examples
For these two examples, make sure you've run the script first.
Simple Example
This just finds the residues b/t chain A and chain B
fetch 1h4g, async=0
interfaceResidues 1h4g
More Complex Example
fetch 1qox, async=0
foundResidues = interfaceResidues("1qox", cA="c. I", cB="c. J", cutoff=0.75, selName="foundIn1QOX")
The Code
from pymol import cmd, stored
def interfaceResidues(cmpx, cA='c. A', cB='c. B', cutoff=1.0, selName="interface"):
"""
interfaceResidues -- finds 'interface' residues between two chains in a complex.
PARAMS
cmpx
The complex containing cA and cB
cA
The first chain in which we search for residues at an interface
with cB
cB
The second chain in which we search for residues at an interface
with cA
cutoff
The difference in area OVER which residues are considered
interface residues. Residues whose dASA from the complex to
a single chain is greater than this cutoff are kept. Zero
keeps all residues.
selName
The name of the selection to return.
RETURNS
* A selection of interface residues is created and named
depending on what you passed into selName
* An array of values is returned where each value is:
( modelName, residueNumber, dASA )
NOTES
If you have two chains that are not from the same PDB that you want
to complex together, use the create command like:
create myComplex, pdb1WithChainA or pdb2withChainX
then pass myComplex to this script like:
interfaceResidues myComlpex, c. A, c. X
This script calculates the area of the complex as a whole. Then,
it separates the two chains that you pass in through the arguments
cA and cB, alone. Once it has this, it calculates the difference
and any residues ABOVE the cutoff are called interface residues.
AUTHOR:
Jason Vertrees, 2009.
"""
# Save user's settings, before setting dot_solvent
oldDS = cmd.get("dot_solvent")
cmd.set("dot_solvent", 1)
# set some string names for temporary objects/selections
tempC, selName1 = "tempComplex", selName+"1"
chA, chB = "chA", "chB"
# operate on a new object & turn off the original
cmd.create(tempC, cmpx)
cmd.disable(cmpx)
# remove cruft and inrrelevant chains
cmd.remove(tempC + " and not (polymer and (%s or %s))" % (cA, cB))
# get the area of the complete complex
cmd.get_area(tempC, load_b=1)
# copy the areas from the loaded b to the q, field.
cmd.alter(tempC, 'q=b')
# extract the two chains and calc. the new area
# note: the q fields are copied to the new objects
# chA and chB
cmd.extract(chA, tempC + " and (" + cA + ")")
cmd.extract(chB, tempC + " and (" + cB + ")")
cmd.get_area(chA, load_b=1)
cmd.get_area(chB, load_b=1)
# update the chain-only objects w/the difference
cmd.alter( "%s or %s" % (chA,chB), "b=b-q" )
# The calculations are done. Now, all we need to
# do is to determine which residues are over the cutoff
# and save them.
stored.r, rVal, seen = [], [], []
cmd.iterate('%s or %s' % (chA, chB), 'stored.r.append((model,resi,b))')
cmd.enable(cmpx)
cmd.select(selName1, 'none')
for (model,resi,diff) in stored.r:
key=resi+"-"+model
if abs(diff)>=float(cutoff):
if key in seen: continue
else: seen.append(key)
rVal.append( (model,resi,diff) )
# expand the selection here; I chose to iterate over stored.r instead of
# creating one large selection b/c if there are too many residues PyMOL
# might crash on a very large selection. This is pretty much guaranteed
# not to kill PyMOL; but, it might take a little longer to run.
cmd.select( selName1, selName1 + " or (%s and i. %s)" % (model,resi))
# this is how you transfer a selection to another object.
cmd.select(selName, cmpx + " in " + selName1)
# clean up after ourselves
cmd.delete(selName1)
cmd.delete(chA)
cmd.delete(chB)
cmd.delete(tempC)
# show the selection
cmd.enable(selName)
# reset users settings
cmd.set("dot_solvent", oldDS)
return rVal
cmd.extend("interfaceResidues", interfaceResidues)