|
|
| (11 intermediate revisions by the same user not shown) |
| Line 1: |
Line 1: |
| == Author == | | {{Infobox script-repo |
| This pymol script is made by Troels Emtekær Linnet
| | |type = script |
| | |filename = displacementmap.py |
| | |author = [[User:Tlinnet|Troels E. Linnet]] |
| | |license = BSD |
| | }} |
|
| |
|
| == Overview == | | == Overview == |
| Line 20: |
Line 24: |
| == Bugs == | | == Bugs == |
| If the criterion is set to low, the memory gets flooded in the data-matrix file, making the file unreadable. No solutions found yet. | | If the criterion is set to low, the memory gets flooded in the data-matrix file, making the file unreadable. No solutions found yet. |
|
| |
| == Instructions ==
| |
| # Make a folder
| |
| # Copy the code to your machine, and name: dispmap.py . You can for example put it into your pymol search path.
| |
| # Download the .pdb files of the Open and Closed form of your protein
| |
| # Make a pymol script file, that makes the alignment and such. See example.
| |
| # Run the script and see the results in command window and suggestions in pymol window
| |
| # Run the gnuplot file to see the data-matrix
| |
|
| |
|
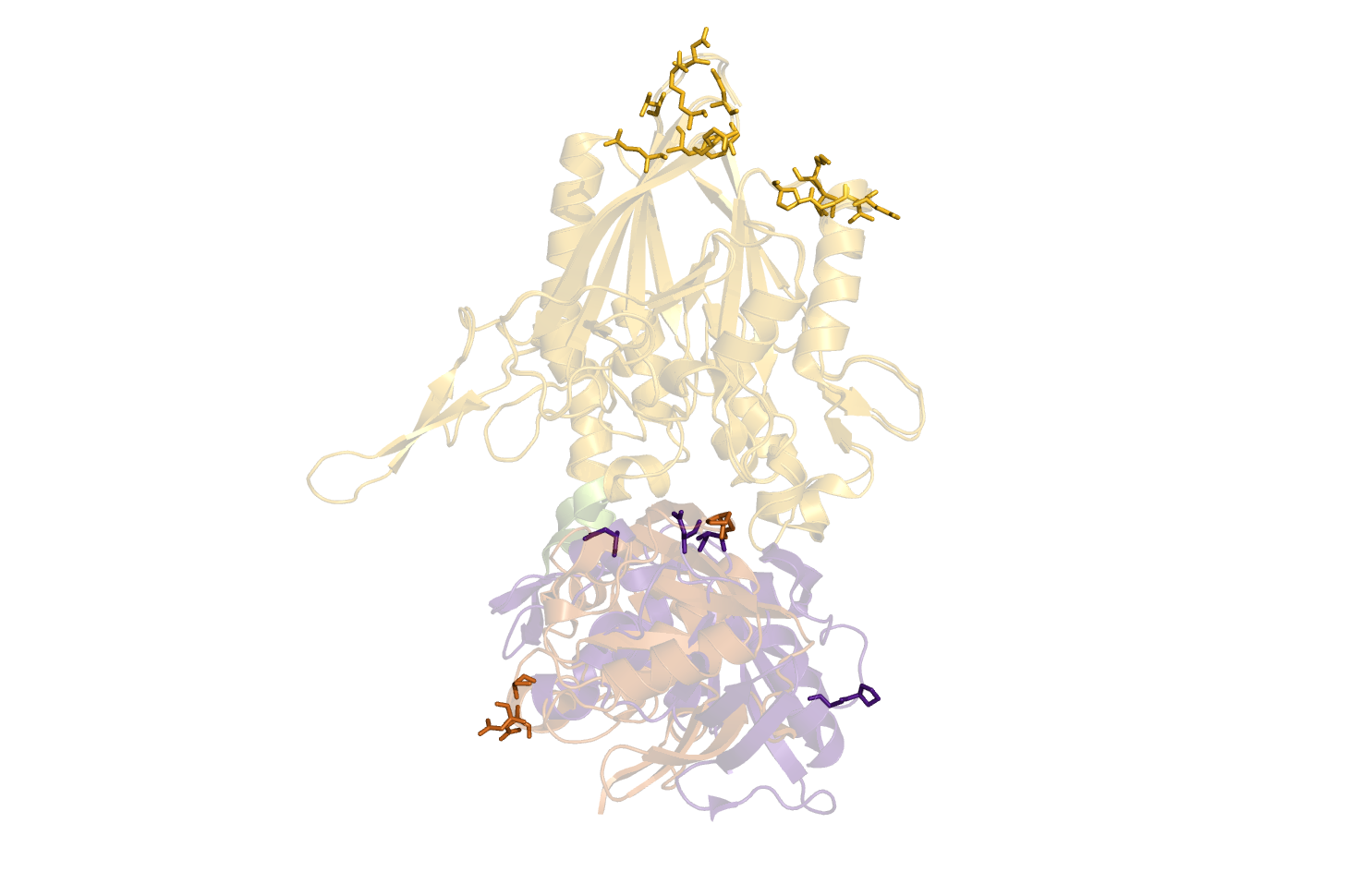
| == Example == | | == Example == |
| Line 51: |
Line 47: |
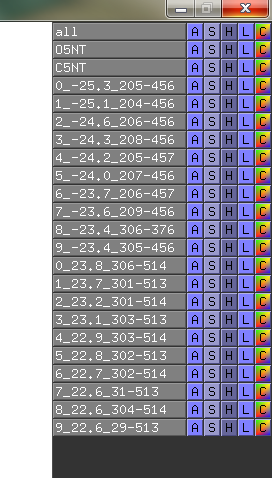
| == Text output == | | == Text output == |
| In the data-matrix.txt file, you find the best suggestions | | In the data-matrix.txt file, you find the best suggestions |
| <source lang="python"> | | <syntaxhighlight lang="text"> |
| # Input 1: Open5NT and Input 2: Closed5NT | | # Input 1: Open5NT and Input 2: Closed5NT |
| # Find for: CA with min. residue-residue dist: 30.0 Angstrom | | # Find for: CA with min. residue-residue dist: 30.0 Angstrom |
| Line 70: |
Line 66: |
| # LEU198 ASP456 -25.3 59.0 33.7 # PRO274 LYS512 24.7 30.3 55.0 # -1/ 1 -3/ 1 # -3/ 2 -1/ 1 # | | # LEU198 ASP456 -25.3 59.0 33.7 # PRO274 LYS512 24.7 30.3 55.0 # -1/ 1 -3/ 1 # -3/ 2 -1/ 1 # |
| # GLN201 ASP456 -25.2 62.8 37.6 # ASN311 PRO513 24.7 35.6 60.3 # -3/ 1 -3/ 1 # -3/ 1 -3/ 2 # | | # GLN201 ASP456 -25.2 62.8 37.6 # ASN311 PRO513 24.7 35.6 60.3 # -3/ 1 -3/ 1 # -3/ 1 -3/ 2 # |
| </source> | | </syntaxhighlight> |
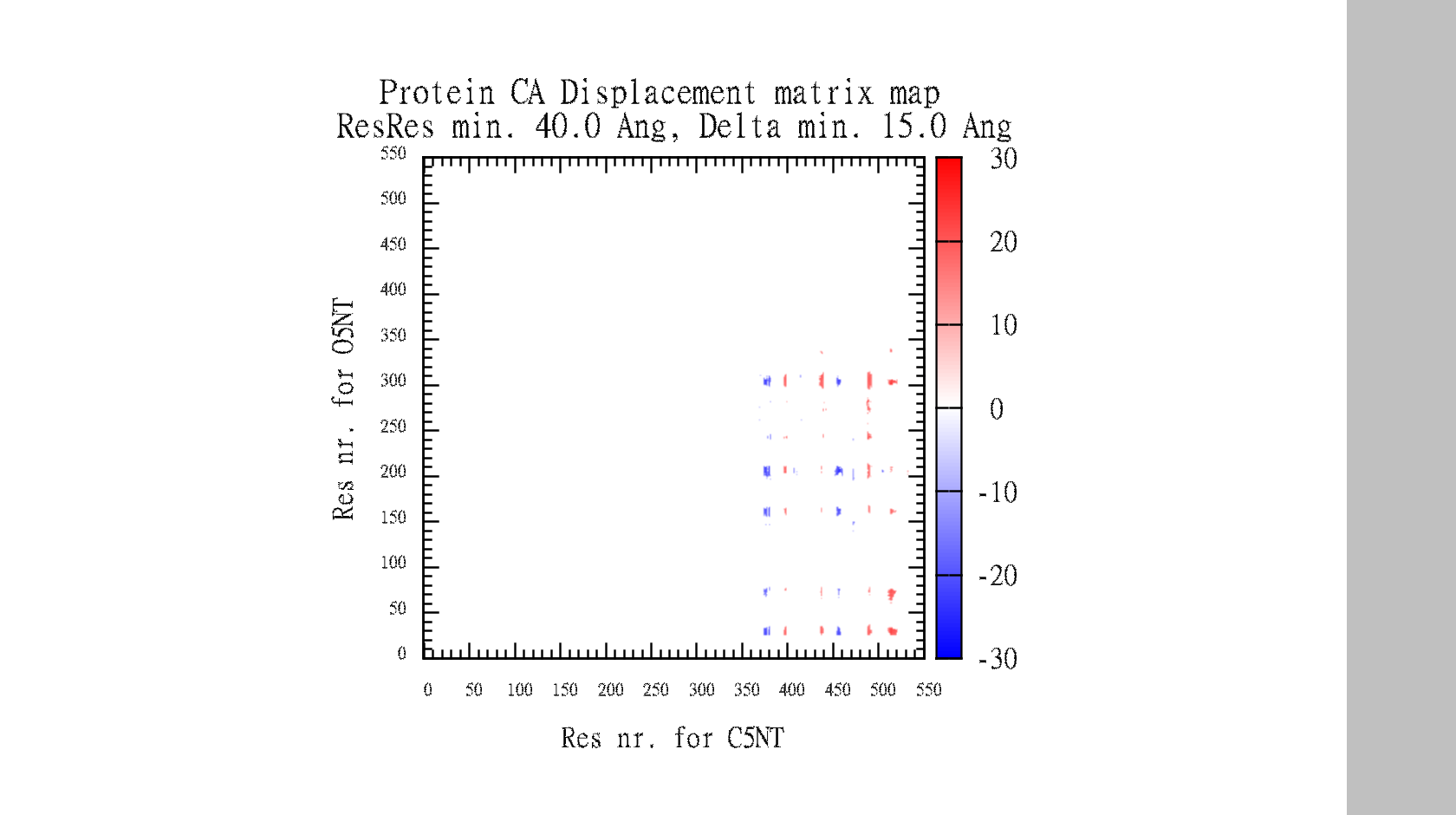
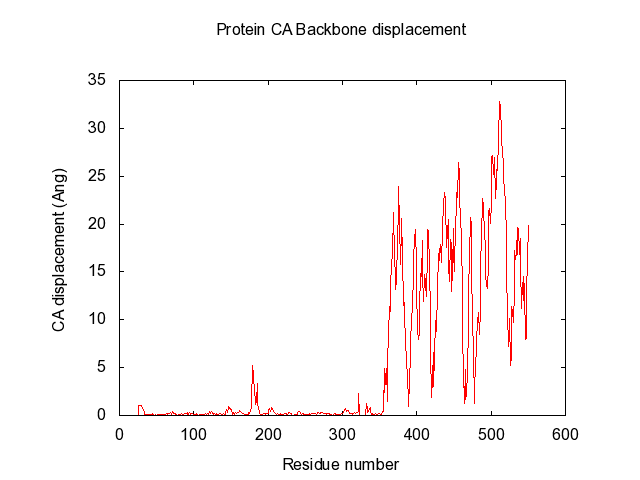
| The script also automatically make the gnuplot plot file (.plt), with all the defined variables, for easy visualisation of the data-matrix.txt and the backbone displacement. | | The script also automatically make the gnuplot plot file (.plt), with all the defined variables, for easy visualisation of the data-matrix.txt and the backbone displacement. |
| <source lang="python">
| |
| reset
| |
| cd '/homes/linnet/Documents/Speciale/5NT-project/Mutant-construct/Distance-Plot/dispmap'
| |
|
| |
| #Title hacks \n is newline, and 0,1 is x,y offset adjustment
| |
| set title "Protein CA Displacement matrix map \n ResRes min. 30.0 Ang, Delta min. 15.0 Ang" 0,1
| |
| # x is column
| |
| set xlabel 'Res nr. for Closed5NT'
| |
| # y is row
| |
| set ylabel 'Res nr. for Open5NT'
| |
|
| |
| #set xrange [300:550]; set yrange [0:400]
| |
| #set xtics 50
| |
| #set ytics 50
| |
| #set mxtics 5
| |
| #set mytics 5
| |
| set size ratio 1
| |
| unset key
| |
|
| |
| set cbrange [-30:30]
| |
| set palette defined (-30 'blue', 0 'white', 30 'red')
| |
| set pm3d map
| |
|
| |
| #set term postscript eps enhanced color
| |
| #set output "Open5NT-Closed5NT-CA-dist.eps"
| |
| set term png
| |
| set output "Open5NT-Closed5NT-CA-dist.png"
| |
| splot 'Open5NT-Closed5NT-CA-dist.txt' matrix
| |
|
| |
| #For the backbone displacement
| |
|
| |
| set title "Protein CA Backbone displacement" 0,1
| |
| set xlabel 'Residue number'
| |
| set ylabel 'CA displacement (Ang)'
| |
|
| |
| #set xrange [0:550]; set yrange [0:40]
| |
| #set xtics 50
| |
| #set ytics 10
| |
| #set mxtics 5
| |
| #set mytics 5
| |
| set size ratio 0.75
| |
| unset key
| |
|
| |
| #set term postscript eps enhanced color
| |
| #set output "Open5NT-Closed5NT-CA-dist-backbone.eps"
| |
| set term png
| |
| set output "Open5NT-Closed5NT-CA-dist-backbone.png"
| |
| plot 'Open5NT-Closed5NT-CA-dist-backbone.txt' using 1:2 title 'Backbone displacement' with lines
| |
| </source>
| |
|
| |
| == Pymol script file ==
| |
| <source lang="python">
| |
| cd /homes/linnet/Documents/Speciale/5NT-project/Mutant-construct/Distance-Plot/dispmap
| |
| #C:\Users\tlinnet\Documents\My Dropbox\Speciale\5NT-project\Mutant-construct\Distance-Plot\dispmap
| |
| ### load pdb files and rename
| |
|
| |
| fetch 1HP1, async=0
| |
| fetch 1HPU, async=0
| |
|
| |
| hide everything, all
| |
| ### Select asymmetric units from pdb file
| |
| create Open5NT, /1HP1//A
| |
| create Closed5NT, /1HPU//A
| |
| delete 1HP1
| |
| delete 1HPU
| |
|
| |
| cartoon auto
| |
| show cartoon, all
| |
| set cartoon_fancy_helices=1
| |
| set bg,[1,1,1]
| |
|
| |
| ### align
| |
| #align Open5NT and resi 26-355, Closed5NT and resi 26-355
| |
| # Super is must faster than align http://www.pymolwiki.org/index.php/Super
| |
| super Open5NT and resi 26-355, Closed5NT and resi 26-355
| |
|
| |
| set auto_zoom, off
| |
| set_view (\
| |
| -0.374262989, 0.692084968, -0.617209554,\
| |
| -0.681849480, -0.656483948, -0.322660774,\
| |
| -0.628496349, 0.300085038, 0.717594206,\
| |
| 0.000000000, 0.000000000, -258.556884766,\
| |
| -0.613845825, 0.472507477, 1.410455704,\
| |
| 205.729583740, 311.384277344, 0.000000000 )
| |
|
| |
| ### Color
| |
| set_color goldenrod1, [1.000, 0.757, 0.145]
| |
| color goldenrod1, resi 26-355
| |
| set_color darkolivegreen1, [0.792, 1.000, 0.439]
| |
| color darkolivegreen1, Open5NT and resi 356-362
| |
| set_color darkolivegreen4, [0.431, 0.545, 0.239]
| |
| color darkolivegreen4, Closed5NT and resi 356-362
| |
| set_color chocolate3, [0.804, 0.400, 0.114]
| |
| color chocolate3, Open5NT and resi 363-550
| |
| set_color purple4, [0.333, 0.102, 0.545]
| |
| color purple4, Closed5NT and resi 363-550
| |
|
| |
| # Select active site
| |
| select active_site, resi 117+120+252+116+217+84+41+43+254
| |
| show sticks, active_site
| |
|
| |
| # Make Cys-Cys bonds
| |
| create SS, (cys/ca+cb+sg) and byres (cys/sg and bound_to cys/sg)
| |
| show sticks, SS
| |
| color yellow, SS
| |
|
| |
| # Mark water molecules
| |
| create waters, resn HOH
| |
| show nb_spheres, waters
| |
| color blue, waters
| |
| disable waters
| |
|
| |
| ### Make sharper, and transparent
| |
| set fog=0
| |
| set cartoon_transparency, 0.7
| |
|
| |
| ### Load the function
| |
| import dispmap
| |
| dispmap
| |
| #dispmap Open5NT, Closed5NT, 40.0, 15.0, resi1=206, resi2=1-512.515-550
| |
| ## Look for serine
| |
| #dispmap mindist=40.0, mindelta=15.0, resi1=206, resi2=330.347.350.405.412.419.457.467.533.534.539.548.336.367.383.397.439.448.490.495.501.518
| |
| #dispmap resi1=308, resi2=513
| |
| </source>
| |
|
| |
| == Python Code ==
| |
| The code can be downloaded fast from here http://tinyurl.com/pymoldispmap <br />
| |
| # wget http://tinyurl.com/pymoldispmap
| |
| # mv pymoldispmap dispmap.py
| |
|
| |
| <source lang="python">
| |
| from pymol import cmd, stored, selector
| |
| from math import *
| |
| import os, re
| |
|
| |
| ## Thx for inspiration from Andreas Henschel
| |
| ## http://www.mail-archive.com/pymol-users@lists.sourceforge.net/msg05595.html (17 dec 2010)
| |
| ## And from Simple scriptin PymMOl http://www.pymolwiki.org/index.php/Simple_Scripting
| |
| ### Ma.Sc student. Troels Linnet, 2010-12-18. troels.linnet@bbz.uni-leipzig.de
| |
|
| |
| ### Calculates the distance for example between all CA atoms between a closed and open form of a structure.
| |
| ### Give a data matrix and a gnuplot file, and input to pymol for easy visualisation
| |
| ### Possible so select interesting residues in ranges. Needs to be separated with a dot '.'
| |
| ### Example input from pymol. with 2 objects.
| |
| ### dispmap O5NT-1HP1-A, C5NT-1HPU-C, 30.0, 15.0, resi1=23-25, atom=CA, showsticks=yes
| |
|
| |
| def dispmap(molecule1="NIL", molecule2="NIL", mindist=30.0, mindelta=15.0, resi1=str(0), resi2=str(0), atom='CA', listlength=5, showsticks='yes'):
| |
| if molecule1=="NIL":
| |
|
| |
| assert len(cmd.get_names())!=0, "Did you forget to load a molecule? There are no objects in pymol."
| |
|
| |
| molecule1=cmd.get_names()[0]
| |
|
| |
| if molecule2=="NIL":
| |
|
| |
| assert len(cmd.get_names())!=0, "Did you forget to load a molecule? There are no objects in pymol."
| |
|
| |
| molecule2=cmd.get_names()[1]
| |
|
| |
| print "You passed in %s and %s" % (molecule1, molecule2)
| |
|
| |
| ### Open filenames
| |
| filename = str(molecule1) + "-" + str(molecule2) + "-" + str(atom) + "-dist"
| |
| backbonefilename = str(molecule1) + "-" + str(molecule2) + "-" + str(atom) + "backbone-dist.txt"
| |
| outfile = open(filename+".txt", "w")
| |
| backboneoutfile = open(filename+"-backbone.txt", "w")
| |
| gnuoutfile = open(filename+".plt", "w")
| |
| print("I have opened matrix %s for you\n" % (filename+".txt"))
| |
|
| |
| ### Sorting for interesting residues for Obj1 and Obj2.
| |
| ### Input is a string, and need to be sorted.
| |
| resi1 = resi1.split('.')
| |
| resi2 = resi2.split('.')
| |
| resi1List = []
| |
| resi2List = []
| |
| for i in resi1:
| |
| if '-' in i:
| |
| tmp = i.split('-')
| |
| resi1List.extend(range(int(tmp[0]),int(tmp[-1])+1))
| |
| if '-' not in i:
| |
| resi1List.append(int(i))
| |
| for i in resi2:
| |
| if '-' in i:
| |
| tmp = i.split('-')
| |
| resi2List.extend(range(int(tmp[0]),int(tmp[-1])+1))
| |
| if '-' not in i:
| |
| resi2List.append(int(i))
| |
| resi1List.sort()
| |
| resi2List.sort()
| |
|
| |
| ### Only take the lines where atom is specified in input
| |
| Object3 = molecule1 + " and name " + str(atom)
| |
| Object4 = molecule2 + " and name " + str(atom)
| |
|
| |
| ### Open 2 name lists
| |
| ### Append residue and atom name to the lists
| |
| stored.OpenPDB = []
| |
| stored.ClosedPDB = []
| |
| cmd.iterate(Object3, "stored.OpenPDB.append((resi, name, resn))")
| |
| cmd.iterate(Object4, "stored.ClosedPDB.append((resi, name, resn))")
| |
|
| |
| ### Open 2 x,y,z position lists
| |
| ### Append atom position
| |
| stored.OpenPos = []
| |
| stored.ClosedPos = []
| |
| cmd.iterate_state(1, selector.process(Object3), "stored.OpenPos.append((x,y,z))")
| |
| cmd.iterate_state(1, selector.process(Object4), "stored.ClosedPos.append((x,y,z))")
| |
|
| |
| ### Sometimes residues gets skipped in X-ray crys, because of low signal or sim. This leads to number conflict.
| |
| ### Make ordered lists according to residue number. Find largest residue number via -1
| |
| OpenOrderedPDB = []
| |
| ClosedOrderedPDB = []
| |
| OpenOrderedPos = []
| |
| ClosedOrderedPos = []
| |
| BackboneDisp = []
| |
|
| |
| ### First fill lists with zeros
| |
| for i in range(int(stored.OpenPDB[-1][0])+1):
| |
| OpenOrderedPDB.append([0,0,0])
| |
| for i in range(int(stored.ClosedPDB[-1][0])+1):
| |
| ClosedOrderedPDB.append([0,0,0])
| |
| for i in range(int(stored.OpenPDB[-1][0])+1):
| |
| OpenOrderedPos.append((0,0,0))
| |
| for i in range(int(stored.ClosedPDB[-1][0])+1):
| |
| ClosedOrderedPos.append((0,0,0))
| |
| for i in range(int(stored.OpenPDB[-1][0])+1):
| |
| BackboneDisp.append([i,0,"NIL",atom])
| |
|
| |
|
| |
| ### Fill in data the right places
| |
| j=0
| |
| for i in stored.OpenPDB:
| |
| OpenOrderedPDB[int(i[0])]=[int(i[0]),i[1],i[2]]
| |
| OpenOrderedPos[int(i[0])]=stored.OpenPos[j]
| |
| j = j + 1
| |
| j=0
| |
| for i in stored.ClosedPDB:
| |
| ClosedOrderedPDB[int(i[0])]=[int(i[0]),i[1],i[2]]
| |
| ClosedOrderedPos[int(i[0])]=stored.ClosedPos[j]
| |
| j = j + 1
| |
|
| |
| ### Make a list with the missing residues
| |
| MissingRes = []
| |
| for index, resi in enumerate(OpenOrderedPDB):
| |
| if abs(OpenOrderedPDB[index][0]-ClosedOrderedPDB[index][0]) != 0:
| |
| MissingRes.append(abs(OpenOrderedPDB[index][0]-ClosedOrderedPDB[index][0]))
| |
| print("Following residues miss in one of the files, and are discarded for")
| |
| print("further calculations")
| |
| print(MissingRes)
| |
| print("")
| |
|
| |
| ### Make the data matrix
| |
| CalcMatrix = create_nXn_matrix(len(OpenOrderedPos))
| |
| print("Calculating a %s X %s distance Matrix" % (len(OpenOrderedPos), len(ClosedOrderedPos)))
| |
|
| |
| ### Make a list with 10 most negative/positive distances
| |
| MaxNegDist = []
| |
| MaxPosDist = []
| |
| for i in range(int(listlength)):
| |
| MaxNegDist.append([0,0,0,0,0,0,0])
| |
| MaxPosDist.append([0,0,0,0,0,0,0])
| |
|
| |
| ### Calculate distances
| |
| for i in range(len(OpenOrderedPos)):
| |
| for j in range(len(ClosedOrderedPos)):
| |
| if OpenOrderedPos[i][0] != 0 and ClosedOrderedPos[j][0] != 0 and OpenOrderedPDB[i][0] not in MissingRes and ClosedOrderedPDB[j][0] not in MissingRes:
| |
| distOpenOpen = distance(OpenOrderedPos, OpenOrderedPos, i, j)
| |
| distClosedClosed = distance(ClosedOrderedPos, ClosedOrderedPos, i, j)
| |
| distOpenClosed = distance(OpenOrderedPos, ClosedOrderedPos, i, j)
| |
| DeltaDist = distOpenClosed - distOpenOpen
| |
| if i == j: BackboneDisp[i] = [i, DeltaDist, OpenOrderedPDB[i][2], atom]
| |
| ###Test if distance is larger than threshold
| |
| if distOpenOpen >= float(mindist) and distClosedClosed >= float(mindist) and abs(DeltaDist) >= float(mindelta):
| |
| CalcMatrix[i][j] = str(round(DeltaDist, 0))
| |
| if DeltaDist < 0 and DeltaDist < MaxNegDist[-1][0] and (i in resi1List or resi1List[-1]==0) and (j in resi2List or resi2List[-1]==0):
| |
| MaxNegDist[-1][0] = DeltaDist
| |
| MaxNegDist[-1][1] = i
| |
| MaxNegDist[-1][2] = j
| |
| MaxNegDist[-1][3] = distOpenOpen
| |
| MaxNegDist[-1][4] = distOpenClosed
| |
| MaxNegDist[-1][5] = str(OpenOrderedPDB[i][2])
| |
| MaxNegDist[-1][6] = str(ClosedOrderedPDB[j][2])
| |
| MaxNegDist = sorted(MaxNegDist)
| |
| if DeltaDist > 0 and DeltaDist > MaxPosDist[-1][0] and (i in resi1List or resi1List[-1]==0) and (j in resi2List or resi2List[-1]==0):
| |
| MaxPosDist[-1][0] = DeltaDist
| |
| MaxPosDist[-1][1] = i
| |
| MaxPosDist[-1][2] = j
| |
| MaxPosDist[-1][3] = distOpenOpen
| |
| MaxPosDist[-1][4] = distOpenClosed
| |
| MaxPosDist[-1][5] = str(OpenOrderedPDB[i][2])
| |
| MaxPosDist[-1][6] = str(ClosedOrderedPDB[j][2])
| |
| MaxPosDist = sorted(MaxPosDist, reverse=True)
| |
|
| |
| print("I made a datamatrix and backbone.txt file for you")
| |
| print("matrix: %s backbone: %s"%(filename+".txt",filename+"-backbone.txt"))
| |
| print("I made a gnuplot file for you, to view the datamatrix and the backbone displacement")
| |
| print("filename: %s\n"%(filename+".plt"))
| |
|
| |
| ###Print distance matrix
| |
| line01="# Input 1: %s and Input 2: %s"%(molecule1,molecule2)
| |
| line02="# Find for: %s with min. residue-residue dist: %s Angstrom"%(atom,mindist)
| |
| line03="# Looking for min. displacement dist: %s Angstrom"%(mindelta)
| |
| line04="# I give nr# suggestions: %s, and do I show sticks in pymol?: %s"%(listlength,showsticks)
| |
| line05="# I look for suggestions in the range: ([0]=>means all residues)"
| |
| line06="# for Input 1: %s and for Input 2: %s"%(resi1, resi2)
| |
| line07="# Mutation info is BLOSUM62 log-odds likelihood score and PAM250 is probability in % for evolutionary distance"
| |
| line08="###########################################################################################################"
| |
| line09="# Max Negative and positive distances # Mutation info #"
| |
| line10="# Obj.1 Obj.2 Delta Op-Op Cl-Cl # Obj.1 Obj.2 Delta Op-Op Cl-Cl # Res.1 Res.2 # Res.1 Res.2 #"
| |
| line11="# Res.1 Res.2 -Dist Dist Dist # Res.1 Res.2 +Dist Dist Dist # B62/PAM250% # B62/PAM250% #"
| |
| outfile.write(line01+'\n')
| |
| outfile.write(line02+'\n')
| |
| outfile.write(line03+'\n')
| |
| outfile.write(line04+'\n')
| |
| outfile.write(line05+'\n')
| |
| outfile.write(line06+'\n')
| |
| outfile.write(line07+'\n')
| |
| outfile.write(line08+'\n')
| |
| outfile.write(line09+'\n')
| |
| outfile.write(line08+'\n')
| |
| outfile.write(line10+'\n')
| |
| outfile.write(line11+'\n')
| |
| outfile.write(line08+'\n')
| |
| print(line01)
| |
| print(line02)
| |
| print(line03)
| |
| print(line04)
| |
| print(line05)
| |
| print(line06)
| |
| print(line07)
| |
| print(line08)
| |
| print(line09)
| |
| print(line08)
| |
| print(line10)
| |
| print(line11)
| |
| print(line08)
| |
| for i in range(len(MaxNegDist)):
| |
| text="# %3s%3s %3s%3s %5s %5s %5s # %3s%3s %3s%3s %5s %5s %5s # %2s/%2s %2s/%2s # %2s/%2s %2s/%2s #"%(MaxNegDist[i][5],MaxNegDist[i][1],MaxNegDist[i][6],MaxNegDist[i][2],round(MaxNegDist[i][0],1),round(MaxNegDist[i][3],1),round(MaxNegDist[i][4],1),MaxPosDist[i][5],MaxPosDist[i][1],MaxPosDist[i][6],MaxPosDist[i][2],round(MaxPosDist[i][0],1),round(MaxPosDist[i][3],1),round(MaxPosDist[i][4],1),cysb62(shortaa(str(MaxNegDist[i][5]))),pam250(shortaa(str(MaxNegDist[i][5]))),cysb62(shortaa(str(MaxNegDist[i][6]))),pam250(shortaa(str(MaxNegDist[i][6]))),cysb62(shortaa(str(MaxPosDist[i][5]))),pam250(shortaa(str(MaxPosDist[i][5]))),cysb62(shortaa(str(MaxPosDist[i][6]))),pam250(shortaa(str(MaxPosDist[i][6]))))
| |
| outfile.write(text+'\n')
| |
| print(text)
| |
| for i in range(len(CalcMatrix)):
| |
| writing = ""
| |
| for j in range(len(CalcMatrix)):
| |
| if str(CalcMatrix[i][j]) == "0.0":
| |
| writing = writing + " " + "?"
| |
| else:
| |
| writing = writing + " " + str(CalcMatrix[i][j])
| |
| ### Add break line
| |
| writing = writing + " " + "\n"
| |
| outfile.write(writing)
| |
| outfile.close()
| |
|
| |
| for i in range(len(BackboneDisp)):
| |
| line="%3s %4s %3s %3s"%(BackboneDisp[i][0],round(BackboneDisp[i][1],1),BackboneDisp[i][2],BackboneDisp[i][3])
| |
| backboneoutfile.write(line+'\n')
| |
| backboneoutfile.close()
| |
|
| |
| ###Make gnuplot plot file
| |
| gnuoutfile.write("reset" + "\n")
| |
| gnuoutfile.write("cd " + "'" + os.getcwd() + "'" + "\n")
| |
| gnuoutfile.write("\n")
| |
| gnuoutfile.write("#Title hacks \\n is newline, and 0,1 is x,y offset adjustment" + "\n")
| |
| gnuoutfile.write('set title "Protein ' + str(atom) + ' Displacement matrix map \\n ResRes min. ' + str(mindist) + ' Ang, ' + 'Delta min. ' + str(mindelta) + ' Ang" 0,1' + "\n")
| |
| gnuoutfile.write("# x is column" + "\n")
| |
| gnuoutfile.write("set xlabel 'Res nr. for " + str(molecule2) +"'"+ "\n")
| |
| gnuoutfile.write("# y is row" + "\n")
| |
| gnuoutfile.write("set ylabel 'Res nr. for " + str(molecule1) +"'"+ "\n")
| |
| gnuoutfile.write("\n")
| |
| gnuoutfile.write("#set xrange [300:550]; set yrange [0:400]" + "\n")
| |
| gnuoutfile.write("#set xtics 50" + "\n")
| |
| gnuoutfile.write("#set ytics 50" + "\n")
| |
| gnuoutfile.write("#set mxtics 5" + "\n")
| |
| gnuoutfile.write("#set mytics 5" + "\n")
| |
| gnuoutfile.write("set size ratio 1" + "\n")
| |
| gnuoutfile.write("unset key" + "\n")
| |
| gnuoutfile.write("\n")
| |
| gnuoutfile.write("set cbrange [-30:30]" + "\n")
| |
| gnuoutfile.write("set palette defined (-30 'blue', 0 'white', 30 'red')" + "\n")
| |
| gnuoutfile.write("set pm3d map" + "\n")
| |
| gnuoutfile.write("\n")
| |
| gnuoutfile.write("#set term postscript eps enhanced color" + "\n")
| |
| gnuoutfile.write('#set output "'+ filename + '.eps"' + "\n")
| |
| gnuoutfile.write("set term png" + "\n")
| |
| gnuoutfile.write('set output "'+ filename + '.png"' + "\n")
| |
| gnuoutfile.write("splot '" + str(filename+".txt") + "' matrix" + "\n")
| |
| gnuoutfile.write("\n")
| |
| gnuoutfile.write("#For the backbone displacement"+"\n")
| |
| gnuoutfile.write("\n")
| |
| gnuoutfile.write('set title "Protein ' + str(atom) + ' Backbone displacement" 0,1' + "\n")
| |
| gnuoutfile.write("set xlabel 'Residue number'" + "\n")
| |
| gnuoutfile.write("set ylabel '" +str(atom) + " displacement (Ang)'" + "\n")
| |
| gnuoutfile.write("\n")
| |
| gnuoutfile.write("#set xrange [0:550]; set yrange [0:40]" + "\n")
| |
| gnuoutfile.write("#set xtics 50" + "\n")
| |
| gnuoutfile.write("#set ytics 10" + "\n")
| |
| gnuoutfile.write("#set mxtics 5" + "\n")
| |
| gnuoutfile.write("#set mytics 5" + "\n")
| |
| gnuoutfile.write("set size ratio 0.75" + "\n")
| |
| gnuoutfile.write("unset key" + "\n")
| |
| gnuoutfile.write("\n")
| |
| gnuoutfile.write("#set term postscript eps enhanced color" + "\n")
| |
| gnuoutfile.write('#set output "'+ filename + '-backbone.eps"' + "\n")
| |
| gnuoutfile.write("set term png" + "\n")
| |
| gnuoutfile.write('set output "'+ filename + '-backbone.png"' + "\n")
| |
| gnuoutfile.write("plot '" + str(filename+"-backbone.txt") + "' using 1:2 title 'Backbone displacement' with lines" + "\n")
| |
| gnuoutfile.close()
| |
|
| |
| ###Create stick residue objects
| |
| for i in range(len(MaxNegDist)):
| |
| name = str(i) + "_" + str(round(MaxNegDist[i][0],1))+"_"+shortaa(str(MaxNegDist[i][5]))+str(MaxNegDist[i][1])+shortaa(str(MaxNegDist[i][6]))+str(MaxNegDist[i][2])
| |
| selection = str(molecule1)+" and resi "+str(MaxNegDist[i][1]) + "+"+str(MaxNegDist[i][2])+" or "+str(molecule2)+" and resi "+str(MaxNegDist[i][2])
| |
| #print selection
| |
| cmd.create(name, selection)
| |
| if showsticks=='yes' or showsticks=='y':
| |
| cmd.show("sticks", name)
| |
| for i in range(len(MaxPosDist)):
| |
| name = str(i) + "_" + str(round(MaxPosDist[i][0],1))+"_"+shortaa(str(MaxPosDist[i][5]))+str(MaxPosDist[i][1])+shortaa(str(MaxPosDist[i][6]))+str(MaxPosDist[i][2])
| |
| selection = str(molecule1)+" and resi " + str(MaxPosDist[i][1])+"+" + str(MaxPosDist[i][2])+" or " + str(molecule2)+" and resi "+str(MaxPosDist[i][2])
| |
| #print selection
| |
| cmd.create(name, selection)
| |
| if showsticks=='yes' or showsticks=='y':
| |
| cmd.show("sticks", name)
| |
| cmd.extend("dispmap",dispmap)
| |
|
| |
| def create_nXn_matrix(n):
| |
| return [[0.0 for x in range(n)] for x in range(n)]
| |
|
| |
| def distance(array1, array2, i, j):
| |
| i = int(i); j = int(j)
| |
| dist = sqrt((array1[i][0] - array2[j][0])**2 + (array1[i][1] - array2[j][1])**2 + (array1[i][2] - array2[j][2])**2)
| |
| return dist
| |
|
| |
| def Coord(Input):
| |
| print cmd.get_atom_coords(Input)
| |
| cmd.extend("Coord",Coord)
| |
|
| |
| def replace_words(text, word_dic):
| |
| rc = re.compile('|'.join(map(re.escape, word_dic)))
| |
| def translate(match):
| |
| return word_dic[match.group(0)]
| |
| return rc.sub(translate, text)
| |
|
| |
| def shortaa(longaa):
| |
| aa_dic = {'ARG':'R','HIS':'H','LYS':'K',
| |
| 'ASP':'D','GLU':'E',
| |
| 'SER':'S','THR':'T','ASN':'N','GLN':'Q',
| |
| 'CYS':'C','SEC':'U','GLY':'G','PRO':'P',
| |
| 'ALA':'A','ILE':'I','LEU':'L','MET':'M','PHE':'F','TRP':'W','TYR':'Y','VAL':'V'}
| |
| return(replace_words(longaa, aa_dic))
| |
|
| |
| def cysb62(aa):
| |
| #BLOSUM62 cys mutation
| |
| # C S T P A G N D E Q H R K M I L V F Y W
| |
| #C9 -1 -1 -3 0 -3 -3 -3 -4 -3 -3 -3 -3 -1 -1 -1 -1 -2 -2 -2
| |
| b62_dic = {'R':'-3','H':'-3','K':'-1',
| |
| 'D':'-3','E':'-4',
| |
| 'S':'-1','T':'-1','N':'-3','Q':'-3',
| |
| 'C':'9','U':'9','G':'-3','P':'-3',
| |
| 'A':'0','I':'-1','L':'-1','M':'-1','F':'-2','W':'-2','Y':'-2','V':'-1'}
| |
| return(replace_words(aa, b62_dic))
| |
|
| |
|
| def pam250(aa):
| | == Example 1 == |
| # A R N D C Q E G H I L K M F P S T W Y V
| | {{Template:PymolScriptRepoDownload|examples/displacementmap_1.pml}} |
| #C 2 1 1 1 52 1 1 2 2 2 1 1 1 1 2 3 2 1 4 2
| | <include src="https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/examples/displacementmap_1.pml" highlight="python" /> |
| # Mutation probability matrix for the evolutionary distance of 250 PAMs.
| |
| # To simplify the appearance, the elements are shown multiplied by 100.
| |
| # In comparing two sequences of average amino acid frequency at this evolutionary distance,
| |
| # there is a 13% probability that a position containing Ala in the first sequence will contain Ala in the second.
| |
| # There is a 3% chance that it will contain Arg, and so forth.
| |
| # (Adapted from Figure 83. Atlas of Protein Sequence and Structure, Suppl 3, 1978, M.O. Dayhoff, ed. National Biomedical Research Foundation, 1979.)
| |
| pam250_dic = {'R':'1','H':'2','K':'1',
| |
| 'D':'1','E':'1',
| |
| 'S':'3','T':'2','N':'1','Q':'1',
| |
| 'C':'52','U':'52','G':'2','P':'2',
| |
| 'A':'2','I':'2','L':'1','M':'1','F':'1','W':'1','Y':'4','V':'2'}
| |
| return(replace_words(aa, pam250_dic))
| |
| </source>
| |
|
| |
|
| == References == | | == References == |
| Line 572: |
Line 95: |
| [[Category:Script_Library|DisplacementMap]] | | [[Category:Script_Library|DisplacementMap]] |
| [[Category:Structural_Biology_Scripts]] | | [[Category:Structural_Biology_Scripts]] |
| | [[Category:Pymol-script-repo]] |