TMalign: Difference between revisions
Jump to navigation
Jump to search
(alignwithanymethod, see also) |
(gallery) |
||
| Line 40: | Line 40: | ||
alignwithanymethod 1C0M, 1BCO | alignwithanymethod 1C0M, 1BCO | ||
</syntaxhighlight> | </syntaxhighlight> | ||
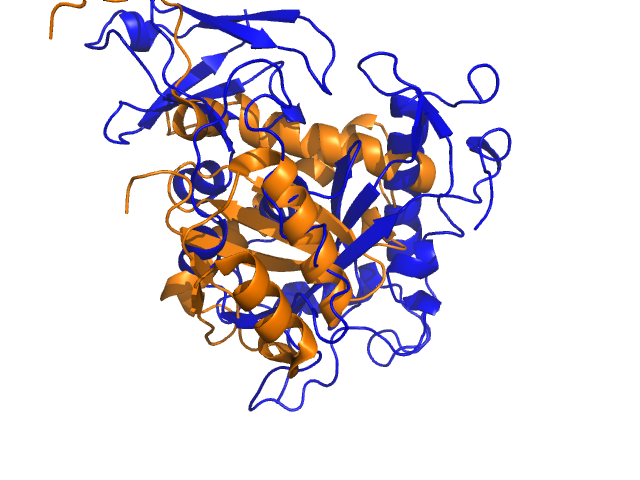
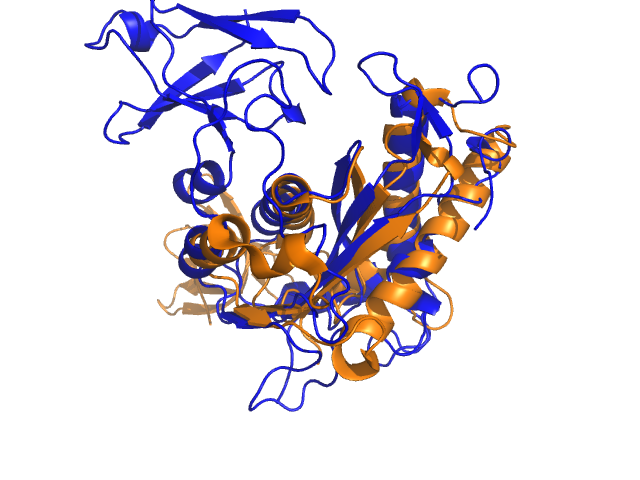
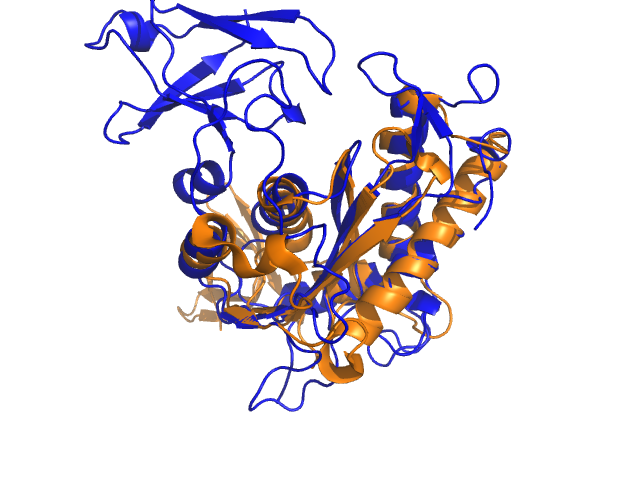
<gallery widths="240" heights="180"> | |||
Image:1C0M_align01.png|1C0M_align01 | |||
Image:1C0M_super01.png|1C0M_super01 | |||
Image:1C0M_cealign01.png|1C0M_cealign01 | |||
Image:1C0M_tmalign01.png|1C0M_tmalign01 | |||
</gallery> | |||
== See Also == | == See Also == | ||
Revision as of 05:15, 14 December 2011
| Type | Python Module |
|---|---|
| Download | tmalign.py |
| Author(s) | Thomas Holder |
| License | BSD |
| This code has been put under version control in the project Pymol-script-repo | |
tmalign is a python module (or script) that provides wrappers to TMalign, TMscore and MMalign. The executables can be downloaded from http://zhanglab.ccmb.med.umich.edu/TM-align/ and should be saved to any directory in PATH.
The module also provides the command alignwithanymethod which is useful to quickly test different alignment methods (with their respective default values).
Usage
tmalign mobile, target [, args [, exe [, ter [, transform [, object ]]]]]
tmscore and mmalign usage is equivalent.
alignwithanymethod mobile, target [, methods ]
Examples
fetch 2xwu 2x19 3gjx, async=0
# TMscore example
tmscore 2x19 and chain B, 2xwu and chain B
# TMalign example with alignment object
tmalign 3gjx and chain A, 2xwu and chain B, object=aln
# full path to executable
mmalign 3gjx, 2x19, exe=/usr/local/src/TM/MMalign
Example for alignwithanymethod (tmalign and cealign will nicely align the beta sheet, but align and super will fail to find a nice superposition):
fetch 1C0M 1BCO, async=0
remove 1C0M and not chain B
alignwithanymethod 1C0M, 1BCO