Nmr cnstr: Difference between revisions
Jump to navigation
Jump to search
m (moved Show NMR constraints to Nmr cnstr: small letters) |
No edit summary |
||
| Line 20: | Line 20: | ||
[[Image:Cns.png]] [[Image:Upl.png]] | [[Image:Cns.png]] [[Image:Upl.png]] | ||
== Python Code == | |||
{{Template:PymolScriptRepoDownload|nmr_cnstr.py}} | |||
[[Category:Script_Library|Show NMR Constraints]] | [[Category:Script_Library|Show NMR Constraints]] | ||
[[Category:Structural_Biology_Scripts]] | [[Category:Structural_Biology_Scripts]] | ||
Revision as of 11:00, 8 December 2011
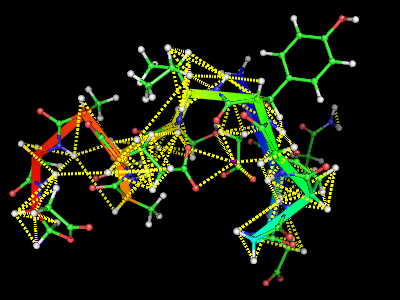
This script will display the NMR constrains used for a structure calculation atop a structure. Usage: Save this as "NMRcnstr.py" load your protein in PyMOL, and run the script. type upl('fname') or cns('fname') where fname is the filename with the NMR constrains you want to display. It is still a very preliminary version.
If you generated the structure by CYANA type:
cyana> read final.pdb
to input the structure in cyana then:
cyana>pseudo=1
before exporting the structure again by:
cyana> write final.pdb
this way the structure will contain the appropriate pseudoatoms nomeclature.
Welcome to contact me if you need some help to set it up.
Python Code
| Download: nmr_cnstr.py | |
| This code has been put under version control in the project Pymol-script-repo | |