Category talk:Script Library
Requests
Please place your PyMOL script requests here. A few other people might have the same request and someone might just code it up for free (viz. all these other scripts). There are no guarantees that someone will write your script, but it doesn't hurt asking.
ExampleScriptTitle
- Example text of what the new awesome script is supposed to do in PyMOL. Consider posting some contact information, so once the script is posted you are apprised.
Instruction request
Would it be possible to add short instructions to category page for new users how to run scripts in pymol...(?)
Policy
Rules
We want the script library to be as valuable as possible to the public. Therefore, please follow these rules when posting code to the wiki:
- Only post code if you have the legal right to do so
- All posted code must be released under some source license or freer (public domain).
- Authors may maintain copyright
- Only post links to code that is not open source licensed, do not copy it onto the wiki
Depositing Scripts
- Create a new page for your script
- Please provide
- an overview of what the script does
- any usage comments or hints
- the source code
- Please add the following to the bottom of your new page:
[[Category:Script_Library]] [[Category:PUT_SUBCATEGORY_NAME_HERE]]
General Notes
Notes
- Feel free to add scripts (see rules below).
- Feel free to add new categories
- You can make script requests on this page's discussion page
- GIT Repository of all our scripts:
git clone git://github.com/jlec/Pymol-script-repo.git
GitHub for Pymol-scripts
I created a github for the pymol scripts. I think this is a good way to to collect and enlarge the collection of scripts. The repo is public accessible so contribute. If you need help on git just write me an email.
http://github.com/jlec/Pymol-script-repo
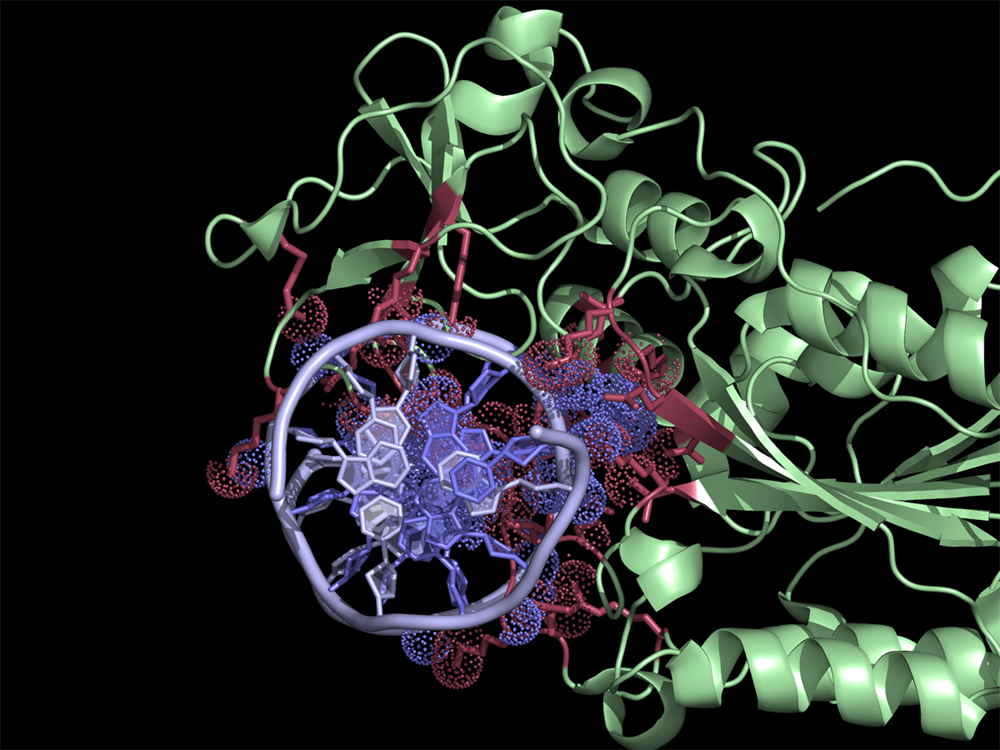
ContactsNCONT
Interface residues (at cutoff <4A) in the 2c7r.pdb were found using NCONT. Usage of ContactsNCONT script in PyMOL allows easy selection of residues and atoms listed in ncont.log file. Interacting protein and DNA residues are colored in red and slate, respectively. Atoms in contact are in dots.
Overview
The script selects residues and atoms from the list of the contacts found by NCONT from CCP4 Program Suite (NCONT analyses contacts between subsets of atoms in a PDB file). First, we run NCONT on our pdb file to find interface residues. Then by using the ContactsNCONT script in PyMOL we select listed residues and atoms separately. This generates two selections (atoms and residues) for each interacting chain, allowing quick manipulation of (sometimes) extensive lists in NCONT log file.
Usage
selectContacts( contactsfile, selName1 = "source", selName2 = "target" )
Examples
First use NCONT to find interface residues/atoms in the same pdb file. Once you have ncont.log file proceed to PyMOL. Make sure you've run the ContactsNCONT script first.
fetch 2c7r selectContacts ncont.log, selName1=prot, selName2=dna
The Code
import re
def parseContacts( f ):
# /1/B/ 282(PHE). / CE1[ C]: /1/E/ 706(GLN). / O [ O]: 3.32
conParser = re.compile("\s*/(\d+)/([A-Z])/\s*(\d+).*?/\s*([A-Z0-9]*).*?:")
mode = 0
s1 = []
s2 = []
pairs = []
for line in f:
if mode == 0:
if line.strip().startswith("SOURCE ATOMS"):
mode = 1
elif mode == 1:
mode = 2
elif mode == 2:
matches = conParser.findall(line)
if len(matches) == 0:
return (s1, s2, pairs)
if len(matches) == 2:
s1.append(matches[0])
s2.append(matches[1])
elif len(matches) == 1:
s2.append(matches[0])
pairs.append((len(s1)-1, len(s2)-1))
else:
print "Unknown mode", mode
def selectContacts( contactsfile, selName1 = "source", selName2 = "target" ):
"""
selectContacts -- parses CCP4 NCONT log file and selects residues and atoms from the list of the contacts found.
PARAMS
contactsfile
filename of the CCP4 NCONT contacts log file
selName1
the name prefix for the _res and _atom selections returned for the
source set of chain
selName2
the name prefix for the _res and _atom selections returned for the
target set of chain
RETURNS
* 2 selections of interface residues and atoms for each chain are created and named
depending on what you passed into selName1 and selName2
AUTHOR:
Gerhard Reitmayr and Dalia Daujotyte, 2009.
"""
# read and parse contacts file into two lists of contact atoms and contact pair list
s1, s2, pairs = parseContacts(open(contactsfile))
# create a selection for the first contact list
resName = selName1 + "_res"
atomName = selName1 + "_atom"
cmd.select(resName, None)
cmd.select(atomName, None)
for (thing, chain, residue, atom) in s1:
cmd.select( resName, resName + " or " + chain+"/"+residue+"/")
cmd.select( atomName, atomName + " or " + chain+"/"+residue+"/"+atom)
# create a selection for the second contact list
resName = selName2 + "_res"
atomName = selName2 + "_atom"
cmd.select(resName, None)
cmd.select(atomName, None)
for (thing, chain, residue, atom) in s2:
cmd.select( resName, resName + " or " + chain+"/"+residue+"/")
cmd.select( atomName, atomName + " or " + chain+"/"+residue+"/"+atom)
cmd.extend("selectContacts", selectContacts)