Symexp
DESCRIPTION
symexp is used to reconstruct neighboring asymmetric units from the crystallographic experiment that produced the given structure. This is assuming the use of a PDB file or equivalent that contains enough information to reproduce the lattice.
USAGE
symexp name_for_new_objects,asymmetric_name,(asymmetric_name),distance
- name_for_new_objects - PyMOL will generate a number of new objects corresponding to copies (rotated and translated) of the given asymmetric unit with the given name appended with a numerical counter
- asymmetric_name - this is the name of the loaded asymmetric unit that you wish to reproduce neighboring crystal partners for
- (asymmetric_name) - the same name, but with parantheses around it - my guess is that the first is the source of atom coordinates and this is the source of the symmetry operators, but that is only a guess.
- distance - in Angstroms; reproduce any other unit that has any part of it falling withing distance Angstroms from the original asymetric unit
EXAMPLE
load any .pdb file into PyMOL (here we use 1GVF).
At the PyMOL command prompt type the following:
>> symexp sym,1GVF,(1GVF),1
produces three new objects. We now have four objects corresponding to two biologic units (the functional protein in a cell).
>> symexp sym,1GVF,(1GVF),5
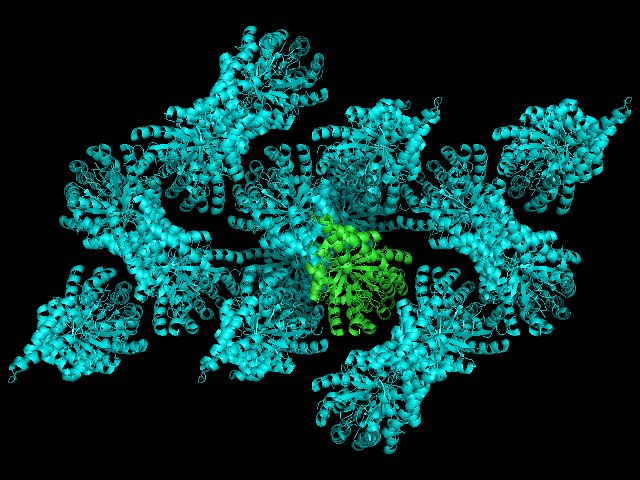
If we color all of the sym* cyan we will produce the following:
As you can see, we can begin to understand the crystal environment of our asymmetric unit. Increasing distance will reveal more of the crystal lattice, but will place in increasing demand on your computer's rendering ability.
NOTE: My PyMOL likes to crash if I ask it to ray trace or make a .png of anything that is too large. I'm not sure how large too large is, but be aware of this.
--Baker1 13:41, 10 November 2008 (CST)