Difference between revisions of "Nmr cnstr"
Jump to navigation
Jump to search
| Line 1: | Line 1: | ||
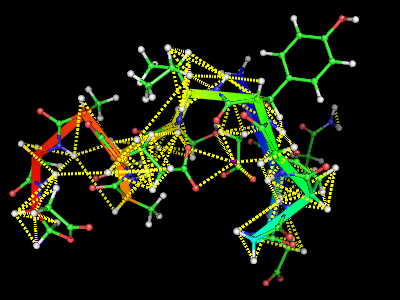
This script will display the NMR constrains used for a structure calculation atop a structure. Usage: Save this as "NMRcnstr.py" load your protein in PyMOL, and run the script. type upl('fname') or cns('fname') where fname is the filename with the NMR constrains you want to display. It is still a very preliminary version. | This script will display the NMR constrains used for a structure calculation atop a structure. Usage: Save this as "NMRcnstr.py" load your protein in PyMOL, and run the script. type upl('fname') or cns('fname') where fname is the filename with the NMR constrains you want to display. It is still a very preliminary version. | ||
| + | |||
| + | If you generate the structure by CYANA type | ||
| + | cyana>pseudo=1 | ||
| + | before exporting the structure by | ||
| + | cyana> write final.pdb | ||
| + | |||
| + | this way the structure will contain the appropriate pseudoatoms nomeclature. | ||
| + | |||
| + | Welcome to contact me if you need some help to set it up. | ||
| + | |||
[[Image:Cns.png]] [[Image:Upl.png]] | [[Image:Cns.png]] [[Image:Upl.png]] | ||
Revision as of 12:55, 19 October 2010
This script will display the NMR constrains used for a structure calculation atop a structure. Usage: Save this as "NMRcnstr.py" load your protein in PyMOL, and run the script. type upl('fname') or cns('fname') where fname is the filename with the NMR constrains you want to display. It is still a very preliminary version.
If you generate the structure by CYANA type cyana>pseudo=1 before exporting the structure by cyana> write final.pdb
this way the structure will contain the appropriate pseudoatoms nomeclature.
Welcome to contact me if you need some help to set it up.
##############################################################################################################
# Pymol Script: For visualizing the NMR constrains (DYANA & CNS format), on top of the calculated structure. #
# Author: Evangelos Papadopoulos. #
# previous affiliation: Dept. of Biochemistry and Biophysics, #
# Arrhenius Laboratories, #
# Stockholm University #
# SE-106 91 Stockholm, Sweden #
# email:evangelos.papadopoulos@gmail.com tel: +468164580 #
# NOTES: This is a preliminary version. #
# #
# '''Reference:''' please if you find this script usefull add the following reference: #
# NMR Solution Structure of the Peptide Fragment 1-30, Derived from Unprocessed Mouse Doppel #
# Protein, in DHPC Micelles. Biochemistry. 2006 Jan 10;45(1):159-166. PMID: 16388591 #
# #
##############################################################################################################
def upl(fname):
f=open(fname,'r')
i=1
upl=f.readline()
#
while upl!='':
print upl,i
cns=string.split(upl)
cmd.dist('upl'+str(i),'i. '+cns[0]+' & n. '+cns[2],'i. '+cns[3]+' & n. '+cns[5])
upl=f.readline()
i+=1
#
f.close()
cmd.hide('labels')
cmd.set ('dash_gap', 0.05)
cmd.do ("orient")
cmd.set ('movie_delay', 1500)
def cns(fname):
f=open(fname,'r')
i=1
upl=f.readline()
print upl,i
while upl!='':
if upl=='\n':
upl=f.readline()
continue
cns=string.split(upl)
print cns,i
if cns[0]=='assign' :
print 'CNS'
if cns[5]=='HB*': print 'CNS***'
cmd.dist('upl'+str(i),'i. '+cns[2]+' & n. '+cns[5],'i. '+cns[7]+' & n. '+cns[10])
i+=1
upl=f.readline()
print '*'+upl+'*',i
f.close()
cmd.set ('dash_gap', 0.05)
cmd.hide('labels')
cmd.do ("orient")
cmd.set ('movie_delay', 1500)