Difference between revisions of "ShowLigandWaters"
Jump to navigation
Jump to search
| Line 39: | Line 39: | ||
output file: waters.py | output file: waters.py | ||
| + | <source lang="python"> | ||
HOH residue 2002 -- and -- ['FAD', '1363', 'O1P'] ---> 2.611289 A | HOH residue 2002 -- and -- ['FAD', '1363', 'O1P'] ---> 2.611289 A | ||
HOH residue 2002 -- and -- ['FAD', '1363', 'O5B'] ---> 3.592691 A | HOH residue 2002 -- and -- ['FAD', '1363', 'O5B'] ---> 3.592691 A | ||
| Line 47: | Line 48: | ||
Number of water molecules that interact with ligand: 3 | Number of water molecules that interact with ligand: 3 | ||
Number of interactions between water molecules and ligand: 4 | Number of interactions between water molecules and ligand: 4 | ||
| + | </source> | ||
[[File:Example 1 FAD 1C0L.png|450px|example #1]] | [[File:Example 1 FAD 1C0L.png|450px|example #1]] | ||
| Line 54: | Line 56: | ||
== The Code == | == The Code == | ||
| − | Copy the following text and save it as | + | Copy the following text and save it as waters.py |
<source lang="python"> | <source lang="python"> | ||
| − | from pymol import cmd, stored, | + | # -*- coding: cp1252 -*- |
| + | """ | ||
| + | This script detects waters molecules within a specified distance from the ligand. | ||
| + | Water molecules are shown. | ||
| + | Distance between water molecules and O or N atoms of ligand are shown. | ||
| + | |||
| + | Usage | ||
| + | waters, [ligand name, distance] | ||
| + | |||
| + | Parameters | ||
| + | |||
| + | ligand : the ligand residue name | ||
| + | |||
| + | distance : a float number that specify the maximum distance from the ligand to consider the water molecule | ||
| + | |||
| + | Output | ||
| + | -A file is produced containing a list of distance between waters and ligand atoms and the number of interactions | ||
| + | -A graphic output share the water molecules interacting whith the ligand atoms by showing the distances between them | ||
| + | """ | ||
| + | |||
| + | from pymol import cmd,stored | ||
| + | #define function: waters | ||
| + | def waters(ligand, distance): | ||
| + | stored.HOH = [] | ||
| + | cmd.set('label_color','white') | ||
| + | cmd.delete ("sele") | ||
| + | cmd.hide ("everything") | ||
| + | cmd.show_as ("sticks", "resn %s" % ligand) | ||
| + | #iterate all water molecules nearby the ligand | ||
| + | cmd.iterate('resn %s around %s and resn HOH' % (ligand,distance),'stored.HOH.append(resi)') | ||
| + | f = open("waters.txt","a") | ||
| + | count=0 | ||
| + | count_int=0 | ||
| + | |||
| + | for i in range(0,len(stored.HOH)): | ||
| + | cmd.distance('dist_HOH_FAD', 'resi ' + stored.HOH[i], '(resn %s and n. O*+N*) w. 3.6 of resi %s'% (ligand, stored.HOH[i])) | ||
| + | stored.name = [] | ||
| + | #iterate all ligand atoms within a predetermined distance from the water molecule | ||
| + | cmd.iterate('(resn %s and n. O*+N*) w. 3.6 of resi %s'% (ligand, stored.HOH[i]),'stored.name.append([resn,resi,name])') | ||
| + | |||
| + | if stored.name: | ||
| + | count = count+1 | ||
| + | count_int = count_int+len(stored.name) | ||
| − | + | for j in range(0,len(stored.name)): | |
| − | + | cmd.select('base', 'resi ' + stored.HOH[i]) | |
| − | + | cmd.select('var','resn '+ligand+ ' and n. ' + stored.name[j][2]) | |
| − | + | #calculate the distance between a specific atom and the water molecule | |
| − | + | dist = cmd.get_distance('base','var') | |
| − | + | f.write('HOH residue %s -- and -- %s ---> %f A\n'%(stored.HOH[i],stored.name[j], dist)) | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | + | cmd.select ("waters","resn %s around %s and resn HOH" % (ligand,distance)) | |
| − | + | cmd.show_as ("spheres", "waters") | |
| − | + | cmd.zoom ("visible") | |
| − | + | num_atm = cmd.count_atoms ("waters") | |
| − | + | print ("Total number of water molecules at %s A from ligand %s: %s \n" % (distance,ligand,num_atm)) | |
| − | + | print ("Number of water molecules that interact with ligand: %d\n" % (count)) | |
| − | + | print ("Number of interactions between water molecules and ligand: %d\n" % count_int) | |
| − | + | f.write('-------------------------------------------------------------\n') | |
| − | + | f.write("Total number of water molecules at %s A from ligand %s: %s \n" % (distance,ligand,num_atm)) | |
| − | + | f.write("Number of water molecules that interact with ligand: %d\n" % count) | |
| − | + | f.write("Number of interactions between water molecules and ligand: %d\n\n\n\n" % count_int) | |
| − | + | f.close() | |
| − | + | cmd.delete ("waters") | |
| − | + | ||
| − | + | cmd.extend("waters",waters) | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
</source> | </source> | ||
Revision as of 05:16, 2 September 2013
| Type | Python Script |
|---|---|
| Download | |
| Author(s) | Gianluca Tomasello |
| License | CC BY-NC-SA |
Introduction
This script detects waters molecules within a specified distance from the ligand. Water molecules are shown. Distance between water molecules and O or N atoms of ligand are shown and is maked an output file containing a list of distance between waters and ligand atoms and the number of interactions
Usage
waters [ligand name, distance]
Required Arguments
- ligand name = the ligand residue name
- distance = max distance in Angstroms
Examples
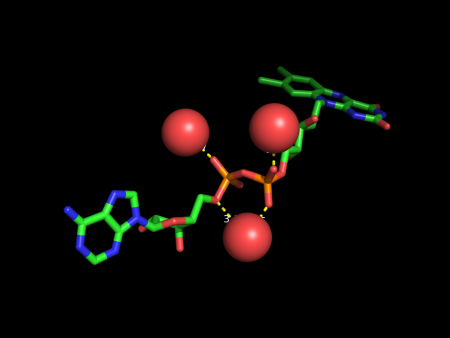
example #1 on PDB structure (1C0L)
PyMOL>waters FAD, 2.8
Total number of water molecules at 2.8 A from ligand FAD: 3
Number of water molecules that interact with ligand: 3
Number of interactions between water molecules and ligand: 4
output file: waters.py
HOH residue 2002 -- and -- ['FAD', '1363', 'O1P'] ---> 2.611289 A
HOH residue 2002 -- and -- ['FAD', '1363', 'O5B'] ---> 3.592691 A
HOH residue 2008 -- and -- ['FAD', '1363', 'O1A'] ---> 2.678604 A
HOH residue 2009 -- and -- ['FAD', '1363', 'O2P'] ---> 2.643039 A
-------------------------------------------------------------
Total number of water molecules at 2.8 A from ligand FAD: 3
Number of water molecules that interact with ligand: 3
Number of interactions between water molecules and ligand: 4
The Code
Copy the following text and save it as waters.py
# -*- coding: cp1252 -*-
"""
This script detects waters molecules within a specified distance from the ligand.
Water molecules are shown.
Distance between water molecules and O or N atoms of ligand are shown.
Usage
waters, [ligand name, distance]
Parameters
ligand : the ligand residue name
distance : a float number that specify the maximum distance from the ligand to consider the water molecule
Output
-A file is produced containing a list of distance between waters and ligand atoms and the number of interactions
-A graphic output share the water molecules interacting whith the ligand atoms by showing the distances between them
"""
from pymol import cmd,stored
#define function: waters
def waters(ligand, distance):
stored.HOH = []
cmd.set('label_color','white')
cmd.delete ("sele")
cmd.hide ("everything")
cmd.show_as ("sticks", "resn %s" % ligand)
#iterate all water molecules nearby the ligand
cmd.iterate('resn %s around %s and resn HOH' % (ligand,distance),'stored.HOH.append(resi)')
f = open("waters.txt","a")
count=0
count_int=0
for i in range(0,len(stored.HOH)):
cmd.distance('dist_HOH_FAD', 'resi ' + stored.HOH[i], '(resn %s and n. O*+N*) w. 3.6 of resi %s'% (ligand, stored.HOH[i]))
stored.name = []
#iterate all ligand atoms within a predetermined distance from the water molecule
cmd.iterate('(resn %s and n. O*+N*) w. 3.6 of resi %s'% (ligand, stored.HOH[i]),'stored.name.append([resn,resi,name])')
if stored.name:
count = count+1
count_int = count_int+len(stored.name)
for j in range(0,len(stored.name)):
cmd.select('base', 'resi ' + stored.HOH[i])
cmd.select('var','resn '+ligand+ ' and n. ' + stored.name[j][2])
#calculate the distance between a specific atom and the water molecule
dist = cmd.get_distance('base','var')
f.write('HOH residue %s -- and -- %s ---> %f A\n'%(stored.HOH[i],stored.name[j], dist))
cmd.select ("waters","resn %s around %s and resn HOH" % (ligand,distance))
cmd.show_as ("spheres", "waters")
cmd.zoom ("visible")
num_atm = cmd.count_atoms ("waters")
print ("Total number of water molecules at %s A from ligand %s: %s \n" % (distance,ligand,num_atm))
print ("Number of water molecules that interact with ligand: %d\n" % (count))
print ("Number of interactions between water molecules and ligand: %d\n" % count_int)
f.write('-------------------------------------------------------------\n')
f.write("Total number of water molecules at %s A from ligand %s: %s \n" % (distance,ligand,num_atm))
f.write("Number of water molecules that interact with ligand: %d\n" % count)
f.write("Number of interactions between water molecules and ligand: %d\n\n\n\n" % count_int)
f.close()
cmd.delete ("waters")
cmd.extend("waters",waters)