Rotamer Toggle
DESCRIPTION
Backbone-Dependent Rotamer library (Dunbrack, Cohen ; see ref) is imported into pymol giving access to this information. There are a number of different ways to use the data, I've only implemented a few as well as added extra functions that seemed useful.
- Rotamer Menu - an added menu into menu.py, which displays the most common rotamers for the given(clicked) residue; you can also set the residue any of the common rotamers as well
- colorRotamers - color rotamers by closest matching rotamer angles from database; i.e. color by how common each rotamer of selection, blue - red (least to most common).
- set_rotamer - routine called by above menu, but can be called manually to set a specific residues side-chain angles
- set_phipsi - set all phi,psi angles of given selection to given angles (useful for creating secondary structures)
- createRotamerPDBs - create pdb for each rotamer of given selection ; filter by rotamer-probability
IMAGES
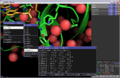
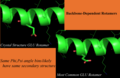
Print out while selecting most common rotamer from above-left image (GLN residue):
Given GLN:40 PHI,PSI (-171.626373291,-96.0500335693) : bin (-170,-100) CHIs: [179.18069458007812, 72.539344787597656, -47.217315673828125] Setting Chi1 to -176.9 Setting Chi2 to 177.4 Setting Chi3 to 0.7
SETUP
run "rotamers.py" and use functions from commandline.
or
To setup a rotamer menu inside the residue menu (default windows pymol installation):
- copy rotamers.py to C:/Program Files/DeLano Scientific/PyMol/modules/pymol/rotamers.py
- copy mymenu.py to C:/Program Files/DeLano Scientific/PyMol/modules/pymol/menu.py (WARNING : overwrites default menu.py - use at your own risk)
- copy bbdep02.May.sortlib to C:/Program Files/DeLano Scientific/PyMol/modules/pymol/bbdep02.May.sortlib (or newer version of sorted bbdep)
This is only one possible way to do this, I am sure there are many others. I'm not going to post the bbdep, but there is a link in the References section to Dunbrack's download page (get the "sorted" lib)
NOTES / STATUS
- Tested on Pymolv0.97, Windows platform, Red Hat Linux 9.0 and Fedora Core 4. Will test v0.98 and MacOSX later on.
- The way it's setup now, when you import rotamers , it will automatically read-in the rotamer database; this may not be what you want.
- Post problems in the discussion page, on 'my talk' page or just email me : dwkulp@mail.med.upenn.edu
TASKS TODO:
- Rotamer Movie, using mset, etc create movie to watch cycle through rotamers
- Code could be organized a bit better; due to time constraints this is good for now..
TASKS DONE:
- Store crystal structure in rotamer menu, so you can go back to original orientation
USAGE
colorRotamers selection set_rotamer selection, chi1_angle [,chi2_angle] [,chi3_angle] [,chi4_angle] set_phipsi selection phi_angle, psi_angle createRotamerPBDs selection [,ncutoff] [,pcutoff] [,prefix]
EXAMPLES
colorRotamers chain A set_rotamer resi 40, -60,-40 (only set chi1,chi2 angles) set_phipsi resi 10-40, -60,-60 (create an alpha-helical-like section) createRotamerPDBs resi 10-12, ncutoff=3 (create 9 PDBs; each with one of the 3 most probable rotamers for resi 10,11,12) createRotamerPDBs resi 14, pcutoff=0.4 (create a pdb file for each rotamer of residue 14 with probablity > 0.4)
REFERENCES
Dunbrack and Cohen. Protein Science 1997
Dunbrack Lab Page (Contains backbone-dependent library)
SCRIPTS (Rotamers.py ; MyMenu.py)
Rotamers.py
##################################################################
# File: Rotamers.py
# Author: Dan Kulp
# Creation Date: 6/8/05
# Contact: dwkulp@mail.med.upenn.edu
#
# Notes:
# Incorporation of Rotamer library
# readRotLib() - fills rotdat;
# indexed by "RES:PHI_BIN:PSI_BIN".
#
# Three main functions:
# 1. colorRotamers - colors according
# to rotamer probablitity
# 2. getBins(sel)
# phi,psi bin for rotamer
# 3. set_rotamer - set a side-chain
# to a specific rotamer
#
# To setup a rotamer menu in the
# right click, under "Residue"
# 1. cp mymenu.py modules/pymol/menu.py
# 2. cp rotamers.py modules/pymol/rotamers.py (update ROTLIB)
#
# Requirements:
# set ROTLIB to path for rotamer library
# Reference:
# Dunbrack and Cohen. Protein Science 1997
####################################################################
import colorsys,sys
import re
import editing
import os
import cmd
import math
# Path for library
ROTLIB=os.environ['PYMOL_PATH']+"/modules/pymol/bbdep02.May.sortlib"
# Place for library in memory..
rotdat = {}
def readRotLib():
# Column indexes in rotamer library..
RES = 0
PHI = 1
PSI = 2
PROB = 8
CHI1 = 9
CHI2 = 10
CHI3 = 11
CHI4 = 12
if os.path.exists(ROTLIB):
print "File exists: "+ROTLIB
input = open(ROTLIB, 'r')
for line in input:
# Parse by whitespace (I believe format is white space and not fixed-width columns)
dat = re.split("\s+",line)
# Add to rotamer library in memory :
# key format RES:PHI_BIN:PSI_BIN
# value format PROB, CHI1, CHI2, CHI3, CHI4
key = dat[RES]+":"+dat[PHI]+":"+dat[PSI]
if key in rotdat:
rotdat[key].append([ dat[PROB], dat[CHI1], dat[CHI2], dat[CHI3], dat[CHI4] ])
else:
rotdat[key] = [ [ dat[PROB], dat[CHI1], dat[CHI2], dat[CHI3], dat[CHI4] ] ]
else:
print "Couldn't find Rotamer library"
# Atoms for each side-chain angle for each residue
CHIS = {}
CHIS["ARG"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","CD" ],
["CB","CG","CD","NE" ],
["CG","CD","NE","CZ" ]
]
CHIS["ASN"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","OD2" ]
]
CHIS["ASP"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","OD1" ]
]
CHIS["CYS"] = [ ["N","CA","CB","SG" ]
]
CHIS["GLN"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","CD" ],
["CB","CG","CD","OE1"]
]
CHIS["GLU"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","CD" ],
["CB","CG","CD","OE1"]
]
CHIS["HIS"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","ND1"]
]
CHIS["ILE"] = [ ["N","CA","CB","CG1" ],
["CA","CB","CG1","CD1" ]
]
CHIS["LEU"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","CD1" ]
]
CHIS["LYS"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","CD" ],
["CB","CG","CD","CE"],
["CG","CD","CE","NZ"]
]
CHIS["MET"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","SD" ],
["CB","CG","SD","CE"]
]
CHIS["PHE"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","CD1" ]
]
CHIS["PRO"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","CD" ]
]
CHIS["SER"] = [ ["N","CA","CB","OG" ]
]
CHIS["THR"] = [ ["N","CA","CB","OG1" ]
]
CHIS["TRP"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","CD1"]
]
CHIS["TYR"] = [ ["N","CA","CB","CG" ],
["CA","CB","CG","CD1" ]
]
CHIS["VAL"] = [ ["N","CA","CB","CG1" ]
]
# Color Rotamer by side-chain angle position
# 'bin' side-chain angles into closest
def colorRotamers(sel):
doRotamers(sel)
# Utility function, to set phi,psi angles for a given selection
# Note: Cartoon, Ribbon functionality will not display correctly after this
def set_phipsi(sel, phi,psi):
doRotamers(sel,angles=[phi,psi],type="set")
# Set a rotamer, based on a selection, a restype and chi angles
def set_rotamer(sel, chi1, chi2=0,chi3=0,chi4=0):
at = cmd.get_model("byres ("+sel+")").atom[0]
list = [chi1,chi2,chi3,chi4]
for i in range(len(CHIS[at.resn])):
print "Setting Chi"+str(i+1)+" to "+str(list[i])
editing.set_dihedral(sel + ' and name '+CHIS[at.resn][i][0],
sel + ' and name '+CHIS[at.resn][i][1],
sel + ' and name '+CHIS[at.resn][i][2],
sel + ' and name '+CHIS[at.resn][i][3], str(list[i]))
# Remove some objects that got created
cmd.delete("pk1")
cmd.delete("pk2")
cmd.delete("pkmol")
# Get Phi,Psi bins for given selection
# WARNING: assume selection is single residue (will only return first residue bins)
def getBins(sel):
return doRotamers(sel, type="bins")
# Color Ramp...
def rot_color(vals):
nbins = 10
vals.sort(key=lambda x: x[1])
# print "End sort: "+str(len(vals))+" : "+str(nbins)
# Coloring scheme...
j = 0
rgb = [0.0,0.0,0.0]
sel_str = ""
for i in range(len(vals)):
if int(len(vals)/nbins) == 0 or i % int(len(vals)/nbins) == 0:
hsv = (colorsys.TWO_THIRD - colorsys.TWO_THIRD * float(j) / (nbins-1), 1.0, 1.0)
#convert to rgb and append to color list
rgb = colorsys.hsv_to_rgb(hsv[0],hsv[1],hsv[2])
if j < nbins-1:
j += 1
cmd.set_color("RotProbColor"+str(i), rgb)
cmd.color("RotProbColor"+str(i), str(vals[i][0]))
# Main function
def doRotamers(sel,angles=[], type="color"):
# Read in Rotamer library if not already done
if len(rotdat) == 0:
readRotLib()
# Set up some variables..
residues = ['dummy'] # Keep track of residues already done
probs = [] # probability of each residue conformation
phi = 0 # phi,psi angles of current residue
psi = 0
# Get atoms from selection
atoms = cmd.get_model("byres ("+sel+")")
# Loop through atoms in selection
for at in atoms.atom:
try:
# Don't process Glycines or Alanines
if not (at.resn == 'GLY' or at.resn == 'ALA'):
if at.chain+":"+at.resn+":"+at.resi not in residues:
residues.append(at.chain+":"+at.resn+":"+at.resi)
# Check for a null chain id (some PDBs contain this)
unit_select = ""
if at.chain != "":
unit_select = "chain "+str(at.chain)+" and "
# Define selections for residue i-1, i and i+1
residue_def = unit_select+'resi '+str(at.resi)
residue_def_prev = unit_select+'resi '+str(int(at.resi)-1)
residue_def_next = unit_select+'resi '+str(int(at.resi)+1)
# Compute phi/psi angle
phi = cmd.get_dihedral(residue_def_prev+' and name C',residue_def+' and name N',residue_def+' and name CA',residue_def+' and name C')
psi = cmd.get_dihedral(residue_def+' and name N',residue_def+' and name CA',residue_def+' and name C',residue_def_next+' and name N')
if type == "set":
print "Changing "+at.resn+str(at.resi)+" from "+str(phi)+","+str(psi)+" to "+str(angles[0])+","+str(angles[1])
cmd.set_dihedral(residue_def_prev+' and name C',residue_def+' and name N',residue_def+' and name CA',residue_def+' and name C',angles[0])
cmd.set_dihedral(residue_def+' and name N',residue_def+' and name CA',residue_def+' and name C',residue_def_next+' and name N', angles[1])
continue
# Find correct 10x10 degree bin
phi_digit = abs(int(phi)) - abs(int(phi/10)*10)
psi_digit = abs(int(psi)) - abs(int(psi/10)*10)
# Remember sign of phi,psi angles
phi_sign = 1
if phi < 0: phi_sign = -1
psi_sign = 1
if psi < 0: psi_sign = -1
# Compute phi,psi bins
phi_bin = int(math.floor(abs(phi/10))*10*phi_sign)
if phi_digit >= 5: phi_bin = int(math.ceil(abs(phi/10))*10*phi_sign)
psi_bin = int(math.floor(abs(psi/10))*10*psi_sign)
if psi_digit >= 5: psi_bin = int(math.ceil(abs(psi/10))*10*psi_sign)
print "Given "+at.resn+":"+at.resi+" PHI,PSI ("+str(phi)+","+str(psi)+") : bin ("+str(phi_bin)+","+str(psi_bin)+")"
# Get current chi angle measurements
chi = []
for i in range(len(CHIS[at.resn])):
chi.append(cmd.get_dihedral(residue_def + ' and name '+CHIS[at.resn][i][0],
residue_def + ' and name '+CHIS[at.resn][i][1],
residue_def + ' and name '+CHIS[at.resn][i][2],
residue_def + ' and name '+CHIS[at.resn][i][3]))
print "CHIs: "+str(chi)
if type == 'bins':
return [at.resn, phi_bin,psi_bin]
# Compute probabilities for given chi angles
prob = 0
prob_box = 22
for item in range(len(rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)])):
print "Rotamer from db: "+str(rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item])
if chi[0]:
if chi[0] >= float(rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][1]) - (prob_box/2) and \
chi[0] <= float(rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][1]) + (prob_box/2):
if len(chi) == 1:
prob = rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][0]
break
if chi[1] >= float(rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][2]) - (prob_box/2) and \
float(chi[1] <= rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][2]) + (prob_box/2):
if len(chi) == 2:
prob = rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][0]
break
if chi[2] >= float(rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][3]) - (prob_box/2) and \
float(chi[2] <= rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][3]) + (prob_box/2):
if len(chi) == 3:
prob = rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][0]
break
if chi[3] >= float(rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][4]) - (prob_box/2) and \
float(chi[3] <= rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][4]) + (prob_box/2):
prob = rotdat[at.resn+":"+str(phi_bin)+":"+str(psi_bin)][item][0]
break
print "PROB OF ROTAMER: "+str(prob)
print "---------------------------"
probs.append([residue_def, prob])
except:
# probs.append([residue_def, -1])
print "Exception found"
continue
# Color according to rotamer probability
rot_color(probs)
# Create PDB files containing most probable rotamers
def createRotamerPDBs(sel,ncutoff=10,pcutoff=0,prefix="ROTAMER"):
# Get atoms from selection
atoms = cmd.get_model("byres ("+sel+")")
# Set up some variables..
residues = ['dummy'] # Keep track of residues already done
# Loop through atoms in selection
for at in atoms.atom:
if at.resn in ('GLY', 'ALA') or "%s:%s:%s" % (at.chain,at.resn,at.resi) in residues:
continue
# Add to residue list (keep track of which ones we've done)
residues.append("%s:%s:%s" % (at.chain,at.resn,at.resi))
# Check for a null chain id (some PDBs contain this)
unit_select = ""
if at.chain != "":
unit_select = "chain "+str(at.chain)+" and "
# Define selections for residue
residue_def = unit_select+'resi '+str(at.resi)
# Get bin (phi,psi) definitions for this residue
bin = doRotamers(residue_def, type='bins')
# Store crystal angle
crystal_angles = [0.0,0.0,0.0,0.0]
for angle in range(3):
try:
crystal_angles[angle] = bin[3][angle]
except IndexError:
break
# Retreive list of rotamers for this phi,psi bin + residue type
match_rotamers = rotdat["%s:%s:%s" % (bin[0],str(bin[1]),str(bin[2]))]
count = 0
for item in range(len(match_rotamers)):
# Store probablity
prob = match_rotamers[item][0]
# Check cutoffs
if float(prob) <= float(pcutoff):
continue
if float(count) >= float(ncutoff):
break
# Increment count
count += 1
# Output to screen ...
print "Residue %s%s, rotamer %i, prob %s" % (at.resn,at.resi,item,prob)
# Set to new rotamer
set_rotamer(residue_def,match_rotamers[item][1],match_rotamers[item][2],match_rotamers[item][3],match_rotamers[item][4])
# Store in PDB file
cmd.save("%s_%s%s_%i_%s.pdb" % (prefix,str(at.resn),str(at.resi),int(item),str(prob)))
# Reset crystal angle
set_rotamer(residue_def,crystal_angles[0],crystal_angles[1],crystal_angles[2],crystal_angles[3])
# Uncommenting this is nice because it loads rotamer library upon startup
# however, it slows the PyMOL loading process a lot
# instead I've put this call into the menuing code..
# readRotLib()
cmd.extend('set_phipsi',set_phipsi)
cmd.extend('set_rotamer',set_rotamer)
cmd.extend('colorRotamers',colorRotamers)
cmd.extend('createRotamerPDBs',createRotamerPDBs)
MyMenu.py
Since menu.py is copyrighted I can't post my edited version, but you can create it very simply by adding these two peices of code
1. In the "pick_option(title,s,object=0)" function of menu.py add the following code after the first "result =" statement
# Edit dwkulp 6/11/05 , add a rotamer menu to residue menu
if title == 'Residue':
result.extend([[ 1, 'rotamers' , rotamer_menu(s)]])
2. At the end of the file add this:
###############################################
# Dan Kulp
# Added Rotamer list to residue menu..
# rotamer.py must be importable (I placed it in
# the same directory as menu.py)
###############################################
import rotamers
def rotamer_menu(s):
# Check for rotamer library being loaded
if not rotamers.rotdat:
rotamers.readRotLib()
# return [ [2, "Must run rotamers.py first",'']]
# Check for valid rotamer residue..
res = cmd.get_model("byres ("+s+")").atom[0].resn
if not res in rotamers.CHIS.keys():
return [ [2, "Residue: "+res+" not known sidechain or does not have rotamers", '']]
# Get PHI,PSI bins for rotamer (also prints out current phi,psi, chi1,chi2,chi3,chi4)
bins = rotamers.doRotamers(s,type='bins')
# Add a title to the menu
result = [ [2, bins[0]+' Rotamers in bin ('+str(bins[1])+','+str(bins[2])+')','' ], [1, ':::PROB,CHI1,CHI2,CHI3,CHI4:::','']]
# Grab the entries for this residue and phi,psi bins
match_rotamers = rotamers.rotdat[bins[0]+":"+str(bins[1])+":"+str(bins[2])]
# Set max number of rotamers to display (probably should be somewhere 'higher up' in the code)
max_rotamers = min(10, len(match_rotamers))
# Create menu entry for each possible rotamer
for item in range(max_rotamers):
result.append( [ 1, str(match_rotamers[item]), 'rotamers.set_rotamer("'+s+'","'\
+str(match_rotamers[item][1])+'","'\
+str(match_rotamers[item][2])+'","'\
+str(match_rotamers[item][3])+'","'\
+str(match_rotamers[item][4])+'")'])
return result