Radius of gyration
Jump to navigation
Jump to search
|
Included in psico | |
| Module | psico.querying#gyradius |
|---|---|
This script does calcualate the radius of gyration of a molecule.
Thanks to Tsjerk Wassenaar for posting this script on the pymol-users mailing list!
The function was added to psico as "gyradius".
from pymol import cmd
import math
def rgyrate(selection='(all)', quiet=1):
'''
DESCRIPTION
Radius of gyration
USAGE
rgyrate [ selection ]
'''
try:
from itertools import izip
except ImportError:
izip = zip
quiet = int(quiet)
model = cmd.get_model(selection).atom
x = [i.coord for i in model]
mass = [i.get_mass() for i in model]
xm = [(m*i,m*j,m*k) for (i,j,k),m in izip(x,mass)]
tmass = sum(mass)
rr = sum(mi*i+mj*j+mk*k for (i,j,k),(mi,mj,mk) in izip(x,xm))
mm = sum((sum(i)/tmass)**2 for i in izip(*xm))
rg = math.sqrt(rr/tmass - mm)
if not quiet:
print("Radius of gyration: %.2f" % (rg))
return rg
cmd.extend("rgyrate", rgyrate)
# vi:expandtab
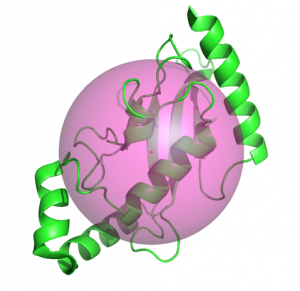
Example
This example uses the radius of gyration and the center of mass to display a semitransparent sphere.
from pymol import cmd, stored, util
import centerOfMass, radiusOfGyration
cmd.set('sphere_transparency', 0.5)
cmd.fetch('2xwu')
cmd.hide('everything')
cmd.show('cartoon', 'chain A')
r = radiusOfGyration.rgyrate('chain A and polymer')
centerOfMass.com('chain A and polymer', object='com', vdw=r)
util.cbc()