The printable version is no longer supported and may have rendering errors. Please update your browser bookmarks and please use the default browser print function instead.
hosted by 
|
Welcome to the PyMOL Wiki!
| The community-run support site for the PyMOL molecular viewer.
|
| To request a new account, email SBGrid at: accounts (@) sbgrid dot org
|
|
|
Did you know...
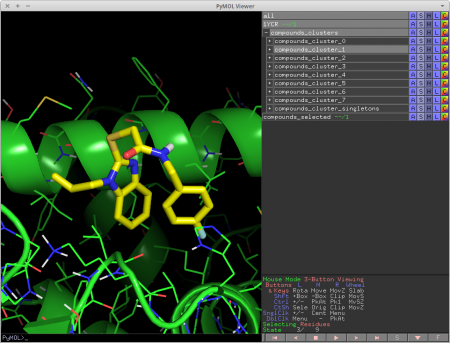
cluster_mols is a PyMOL plugin that allows the user to quickly select compounds from a virtual screen to be purchased or synthesized.
It helps the user by automatically clustering input compounds based on their molecular fingerprints [1] and loading them into the PyMOL window. cluster_mols also highlights both good and bad polar interactions between the ligands and a user specified receptor. Additionally there are a number of keyboard controls for selecting and extracting compounds, as well as functionality for searching online to see if there are vendors for a selected compound.
Description
The basic work flow of cluster_mols.py can be broken up into three parts.
- Computing a similarity matrix from the input compounds
- Performing hierarchical clustering on the results from 1)
- Cutting the tree at a user-specified height and creating and sorting clusters
The results of 1 and 2 are saved to python pickle files so you do not have to recompute them in ..→
|
|
|
 A Random PyMOL-generated Cover. See Covers.
|