Difference between revisions of "MOLE 2.0: advanced approach for analysis of biomacromolecular channels"
LukasPravda (talk | contribs) m (wrong url corrected) |
LukasPravda (talk | contribs) |
||
| Line 1: | Line 1: | ||
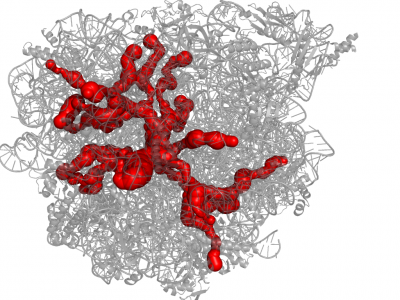
| − | [[File:Molelogo.png|80px|left|]] [[File:1jj2.png|400px|thumb|right|Example of channel system of 1JJ2]] [http://mole.chemi.muni.cz MOLE 2 | + | [[File:Molelogo.png|80px|left|]] [[File:1jj2.png|400px|thumb|right|Example of channel system of 1JJ2]] [http://mole.chemi.muni.cz MOLE 2] is a successor of a popular software tool [http://lcc.ncbr.muni.cz/whitezone/development/mole/web/index.php Mole] for detection and characterization of channels in biomacromolecules (proteins, nucleic acids and glycans). This completely redesigned version allows user rapid and accurate '''analysis of channels and transmembrane pores''' even ''' in large structures''' (hundreds of thousands of atoms). As a new feature, MOLE 2 '''estimates''' not only '''physicochemical properties''' of the identified channels, i.e., '''hydropathy, hydrophobicity, polarity, charge, and mutability''', but also physicochemical properties of the cavities. For thorough description of the functionality and instructions on working with MOLE 2 please consult the [http://dx.doi.org/10.1186/1758-2946-5-39 paper] or our [http://webchem.ncbr.muni.cz/Platform/AppsBin/Mole2_manual.pdf manual]. |
| + | |||
| + | ==MOLE 2.5 Update== | ||
| + | Update of MOLE 2 with novel functionality and bug fixes has been released. The binaries and plugins are available for download from the [https://webchem.ncbr.muni.cz/Platform/App/Mole | main page]. | ||
| + | * Easily remove parts of the PDB entry with [https://webchem.ncbr.muni.cz/Wiki/PatternQuery:UserManual PatternQuery]. | ||
| + | * New PDB standard mmCIF is supported and recommended for all calculations. | ||
| + | * New weight functions for tunnel/pore calculation. | ||
==Availability and Requirements== | ==Availability and Requirements== | ||
| − | MOLE 2. | + | MOLE 2.5 is available as a [https://webchem.ncbr.muni.cz/Platform/App/Mole GUI application] with in-built molecular browser enabling user interactive work; [https://webchem.ncbr.muni.cz/Platform/App/Mole command-line application] and [https://webchem.ncbr.muni.cz/Platform/App/Mole PyMOL & Chimera plugin]. Some functionality is also available via online [http://beta.mole.upol.cz web service]. |
* GUI application is available for Windows with [http://www.microsoft.com/en-us/download/details.aspx?id=17851 .NET 4.0 framework] or newer installed. | * GUI application is available for Windows with [http://www.microsoft.com/en-us/download/details.aspx?id=17851 .NET 4.0 framework] or newer installed. | ||
| − | * Command-line application and PyMOL plugin are available for Windows, Linux (Unix) and MacOS. For Linux (Unix) and MacOS systems [http://mono-project.com/Main_Page Mono framework] 2.10 or newer. | + | * Command-line application and PyMOL&Chimera plugin are available for Windows, Linux (Unix) and MacOS. For Linux (Unix) and MacOS systems [http://mono-project.com/Main_Page Mono framework] 2.10 or newer is required. |
| − | |||
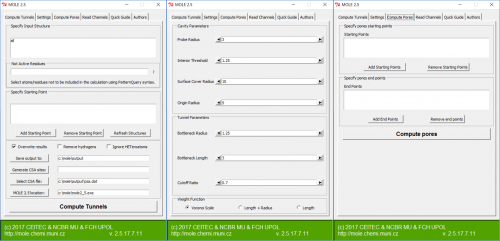
| − | [[File:Plugin.png|500px|thumb|center|MOLE 2. | + | [[File:Plugin.png|500px|thumb|center|MOLE 2.5 PyMOL plugin]] |
==References== | ==References== | ||
| − | # Sehnal D, Svobodová Vařeková R, Berka K, Pravda L, Navrátilová V, Banáš P, Ionescu C-M, Otyepka M, Koča J: [http://dx.doi.org/10.1186/1758-2946-5-39 MOLE 2.0: advanced approach for analysis of biomacromolecular channels. Journal of Cheminformatics 2013, 5:39.] | + | # Sehnal D, Svobodová Vařeková R, Berka K, Pravda L, Navrátilová V, Banáš P, Ionescu C-M, Otyepka M, Koča J: [http://dx.doi.org/10.1186/1758-2946-5-39 MOLE 2.0: advanced approach for analysis of biomacromolecular channels]. Journal of Cheminformatics 2013, 5:39. |
| − | # Berka K, Hanák O, Sehnal D, Banáš P, Navrátilová V, Jaiswal D, Ionescu C-M, Svobodová Vařeková R, Koča J, Otyepka M: [http://dx.doi.org/10.1093/nar/GKS363 MOLEonline 2.0: interactive web-based analysis of biomacromolecular channels. Nucleic acids research 2012, 40:W222–7. | + | # Pravda,L., Berka,K., Svobodová Vařeková,R., Sehnal,D., Banáš,P., Laskowski,R.A., Koča,J. and Otyepka,M. (2014) [http://dx.doi.org/10.1186/s12859-014-0379-x Anatomy of enzyme channels]. BMC Bioinformatics, 15, 379. |
| + | # Berka K, Hanák O, Sehnal D, Banáš P, Navrátilová V, Jaiswal D, Ionescu C-M, Svobodová Vařeková R, Koča J, Otyepka M: [http://dx.doi.org/10.1093/nar/GKS363 MOLEonline 2.0: interactive web-based analysis of biomacromolecular channels]. Nucleic acids research 2012, 40:W222–7. | ||
[[Category:Surfaces and Voids|Mole]] | [[Category:Surfaces and Voids|Mole]] | ||
Latest revision as of 07:38, 13 July 2017
MOLE 2 is a successor of a popular software tool Mole for detection and characterization of channels in biomacromolecules (proteins, nucleic acids and glycans). This completely redesigned version allows user rapid and accurate analysis of channels and transmembrane pores even in large structures (hundreds of thousands of atoms). As a new feature, MOLE 2 estimates not only physicochemical properties of the identified channels, i.e., hydropathy, hydrophobicity, polarity, charge, and mutability, but also physicochemical properties of the cavities. For thorough description of the functionality and instructions on working with MOLE 2 please consult the paper or our manual.
MOLE 2.5 Update
Update of MOLE 2 with novel functionality and bug fixes has been released. The binaries and plugins are available for download from the | main page.
- Easily remove parts of the PDB entry with PatternQuery.
- New PDB standard mmCIF is supported and recommended for all calculations.
- New weight functions for tunnel/pore calculation.
Availability and Requirements
MOLE 2.5 is available as a GUI application with in-built molecular browser enabling user interactive work; command-line application and PyMOL & Chimera plugin. Some functionality is also available via online web service.
- GUI application is available for Windows with .NET 4.0 framework or newer installed.
- Command-line application and PyMOL&Chimera plugin are available for Windows, Linux (Unix) and MacOS. For Linux (Unix) and MacOS systems Mono framework 2.10 or newer is required.
References
- Sehnal D, Svobodová Vařeková R, Berka K, Pravda L, Navrátilová V, Banáš P, Ionescu C-M, Otyepka M, Koča J: MOLE 2.0: advanced approach for analysis of biomacromolecular channels. Journal of Cheminformatics 2013, 5:39.
- Pravda,L., Berka,K., Svobodová Vařeková,R., Sehnal,D., Banáš,P., Laskowski,R.A., Koča,J. and Otyepka,M. (2014) Anatomy of enzyme channels. BMC Bioinformatics, 15, 379.
- Berka K, Hanák O, Sehnal D, Banáš P, Navrátilová V, Jaiswal D, Ionescu C-M, Svobodová Vařeková R, Koča J, Otyepka M: MOLEonline 2.0: interactive web-based analysis of biomacromolecular channels. Nucleic acids research 2012, 40:W222–7.