Difference between revisions of "Load new B-factors"
Jump to navigation
Jump to search
(Page created) |
m (download link added) |
||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = script | |type = script | ||
| − | |download = | + | |download = [http://dx.doi.org/10.6084/m9.figshare.1176991 figshare] |
|author = [[User:PietroGattiLafranconi|Pietro Gatti-Lafranconi]] | |author = [[User:PietroGattiLafranconi|Pietro Gatti-Lafranconi]] | ||
|license = [http://creativecommons.org/licenses/by/4.0/ CC BY 4.0] | |license = [http://creativecommons.org/licenses/by/4.0/ CC BY 4.0] | ||
Latest revision as of 11:43, 22 September 2014
| Type | Python Script |
|---|---|
| Download | figshare |
| Author(s) | Pietro Gatti-Lafranconi |
| License | CC BY 4.0 |
Introduction
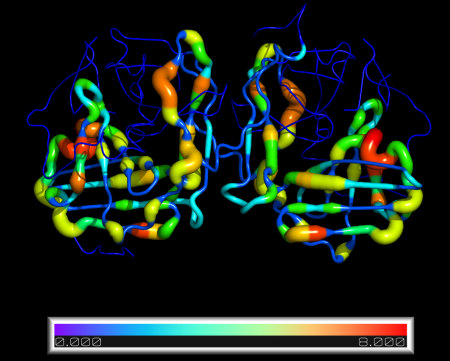
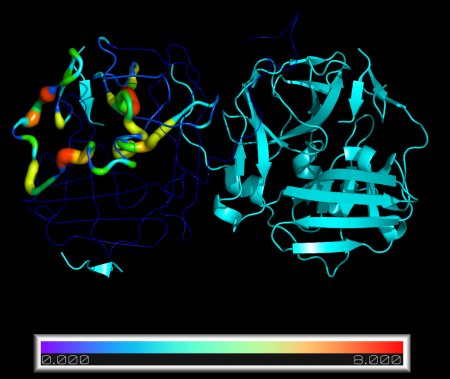
Quick and simple script to replace B-factor values in a PDB structure.
New B-factors are provided in an external .txt file, one per line.
By default, the script will also redraw the selected molecule as cartoon putty and colour by B-factor
Usage
loadBfacts mol, [startaa, [source, [visual Y/N]]]
Required Arguments
- mol = any object selection (within one single object though)
Optional Arguments
- startaa = number of amino acid the first value in 'source' refers to (default=1)
- source = name of the file containing new B-factor values (default=newBfactors.txt)
- visual = redraws structure as cartoon_putty and displays bar with min/max values (default=Y)
Limitations
For its very nature, this script is not suitable for complex cases in which only some B-factors are to be replaced, or on complex selections. In such cases, data2bfactor.py is a better choice.
B-factors have to be entered one per line, with no labels or amino acid ID. They will replace values of a continuous stretch of amino acids of equal length (or shorter) starting from the position provided with 'startaa'.
'newBfactors.txt' looks like
2 0 0 3 7 ...
Examples
PyMOL>loadbfacts 1LVM
PyMOL>loadbfacts 1LVM and chain a, startaa=135
The Code
from pymol import cmd, stored, math
def loadBfacts (mol,startaa=1,source="newBfactors.txt", visual="Y"):
"""
Replaces B-factors with a list of values contained in a plain txt file
usage: loadBfacts mol, [startaa, [source, [visual]]]
mol = any object selection (within one single object though)
startaa = number of first amino acid in 'new B-factors' file (default=1)
source = name of the file containing new B-factor values (default=newBfactors.txt)
visual = redraws structure as cartoon_putty and displays bar with min/max values (default=Y)
example: loadBfacts 1LVM and chain A
"""
obj=cmd.get_object_list(mol)[0]
cmd.alter(mol,"b=-1.0")
inFile = open(source, 'r')
counter=int(startaa)
bfacts=[]
for line in inFile.readlines():
bfact=float(line)
bfacts.append(bfact)
cmd.alter("%s and resi %s and n. CA"%(mol,counter), "b=%s"%bfact)
counter=counter+1
if visual=="Y":
cmd.show_as("cartoon",mol)
cmd.cartoon("putty", mol)
cmd.set("cartoon_putty_scale_min", min(bfacts),obj)
cmd.set("cartoon_putty_scale_max", max(bfacts),obj)
cmd.set("cartoon_putty_transform", 0,obj)
cmd.set("cartoon_putty_radius", 0.2,obj)
cmd.spectrum("b","rainbow", "%s and n. CA " %mol)
cmd.ramp_new("count", obj, [min(bfacts), max(bfacts)], "rainbow")
cmd.recolor()
cmd.extend("loadBfacts", loadBfacts);