Label
DESCRIPTION
label labels one or more atoms properties over a selection using the python evaluator with a separate name space for each atom. The symbols defined in the name space are:
- name
- resn
- resi
- chain
- q
- b
- segi
- type (ATOM,HETATM)
- formal_charge
- partial_charge
- numeric_type
- text_type
All strings in the expression must be explicitly quoted. This operation typically takes several seconds per thousand atoms altered. To clear labels, simply omit the expression or set it to .
USAGE
label (selection),expression
SETTINGS
FONT
There are 9 different scalable fonts.
set label_font_id, number
where number is 5 through 14.
SIZE
The font size can be adjusted
set label_size, number
where number is the point size (or -number for Angstroms)
COLOR
Set a label's color by
set label_color, color
where color is a valid PyMol color.
EXPRESSION
To set what the label reads (see above)
label selection, expression
For example
label all, name
label resi 10, b
POSITION
To position labels
edit_mode
then ctrl-middle-click-and-drag to position the label in space.
EXAMPLES
label (chain A),chain label (n;ca),"%s-%s" % (resn,resi) label (resi 200),"%1.3f" % partial_charge
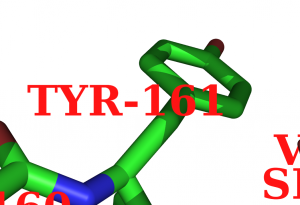
The following image was created with
label (resi 200),"%1.3f" % b
set label_font_id, 10
set label_size, 10
and finally, some labels were moved around in edit_mode.
Users Comments
Labels Using ID Numbers
The following commnent,
label SELECTION, " %s" % ID
labels the SELECTION with atom ID numbers.
You can make more complicated selections/lables such as
label SELECTION, " %s:%s %s" % (resi, resn, name)
which will give you something like "GLU:139 CG"
Lables Using Three Letter Abbreviations
- First, Add this to your $HOME/.pymolrc file:
# start $HOME/.pymolrc modification
one_letter =3D {'VAL':'V', 'ILE':'I', 'LEU':'L', 'GLU':'E', 'GLN':'Q', \
'ASP':'D', 'ASN':'N', 'HIS':'H', 'TRP':'W', 'PHE':'F', 'TYR':'Y', \
'ARG':'R', 'LYS':'K', 'SER':'S', 'THR':'T', 'MET':'M', 'ALA':'A', \
'GLY':'G', 'PRO':'P', 'CYS':'C'}
# end modification
- . Second, instead of:
label n. ca, resn
use:
label n. ca, one_letter[resn]