Difference between revisions of "Inertia tensor"
Jump to navigation
Jump to search
m (→Reference) |
|||
| (3 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
|type = Python Module | |type = Python Module | ||
|filename = inertia_tensor.py | |filename = inertia_tensor.py | ||
| − | |author = [ | + | |author = [http://www.mattmaciejewski.com Mateusz Maciejewski] |
|license = MIT | |license = MIT | ||
}} | }} | ||
| + | |||
| + | [[Image:It.png|300px|right]] | ||
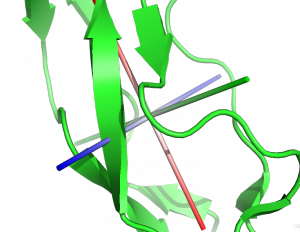
This script will draw the eigenvectors of the inertia tensor of the selected part of the molecule. | This script will draw the eigenvectors of the inertia tensor of the selected part of the molecule. | ||
| Line 17: | Line 19: | ||
<source lang="python"> | <source lang="python"> | ||
| − | fetch 4b2r | + | fetch 4b2r, async=0 |
hide lines | hide lines | ||
show cartoon | show cartoon | ||
run inertia_tensor.py | run inertia_tensor.py | ||
| − | tensor 4b2r & n. n+ca+c & i. 569- | + | tensor 4b2r & n. n+ca+c & i. 569-623, tensor_dom11, 1 |
</source> | </source> | ||
Latest revision as of 21:55, 7 March 2013
| Type | Python Module |
|---|---|
| Download | inertia_tensor.py |
| Author(s) | Mateusz Maciejewski |
| License | MIT |
| This code has been put under version control in the project Pymol-script-repo | |
This script will draw the eigenvectors of the inertia tensor of the selected part of the molecule.
Usage
tensor selection, name, model
Examples
To draw the inertia tensor of the first model in PDB file 4B2R do:
fetch 4b2r, async=0
hide lines
show cartoon
run inertia_tensor.py
tensor 4b2r & n. n+ca+c & i. 569-623, tensor_dom11, 1
Reference
A figure generated using this script can be seen in the following reference:
- Estimation of interdomain flexibility of N-terminus of factor H using residual dipolar couplings. Maciejewski M, Tjandra N, Barlow PN. Biochemistry. 50(38) 2011, p. 8138-49, fig. 1 doi:10.1021/bi200575b