Difference between revisions of "Huge surfaces"
Jump to navigation
Jump to search
(gaussian_b_floor) |
(CA-only) |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
PyMOL can render '''very large''' (huge) surfaces of proteins, using a few tricks. First off, you should know that because of the size, you do not get the level of accuracy that you would for a smaller molecule. However, if you need to represent a molecule (or object) with a few million atoms, this may be your option of choice. | PyMOL can render '''very large''' (huge) surfaces of proteins, using a few tricks. First off, you should know that because of the size, you do not get the level of accuracy that you would for a smaller molecule. However, if you need to represent a molecule (or object) with a few million atoms, this may be your option of choice. | ||
| Line 10: | Line 9: | ||
</gallery> | </gallery> | ||
| − | = | + | == Strategy 1: Low resolution Gaussian Map == |
<source lang="python"> | <source lang="python"> | ||
# load a whopping big PDB | # load a whopping big PDB | ||
| − | fetch 1aon, struct | + | fetch 1aon, struct, async=0 |
# create a color spectrum over the object | # create a color spectrum over the object | ||
spectrum count, selection=struct | spectrum count, selection=struct | ||
| − | |||
| − | |||
| − | |||
| − | |||
# === now create a pseudo-fcalc map (a 3D volumetric scalar field) === | # === now create a pseudo-fcalc map (a 3D volumetric scalar field) === | ||
# set the B-factors nice and high for smoothness | # set the B-factors nice and high for smoothness | ||
| − | + | set gaussian_b_floor, 40 | |
# ~10 A map resolution | # ~10 A map resolution | ||
set gaussian_resolution, 10 | set gaussian_resolution, 10 | ||
| − | |||
| − | |||
| − | |||
# ~10 A map spacing with a 10 A surrounding buffer | # ~10 A map spacing with a 10 A surrounding buffer | ||
| Line 40: | Line 32: | ||
# now color the map based on the underlying protein | # now color the map based on the underlying protein | ||
| − | ramp_new ramp, struct, [0 | + | ramp_new ramp, struct, [0,10], [atomic, atomic] |
color ramp, surf | color ramp, surf | ||
| Line 46: | Line 38: | ||
# hide the ramp | # hide the ramp | ||
disable ramp | disable ramp | ||
| + | </source> | ||
| + | |||
| + | == Strategy 2: CA-only model with oversized radii == | ||
| + | <source lang="python"> | ||
| + | # load a whopping big PDB | ||
| + | fetch 1aon, struct, async=0 | ||
| + | |||
| + | # reduce to CA atoms only | ||
| + | remove struct & not guide | ||
| + | |||
| + | # create a color spectrum over the object | ||
| + | spectrum count, selection=struct | ||
| + | |||
| + | # increase VDW radii to compensate volume of missing non-CA atoms | ||
| + | alter struct, vdw=4 | ||
| + | |||
| + | # increase VDW radius of solvent | ||
| + | set solvent_radius, 4, struct | ||
| + | |||
| + | # show molecular surface | ||
| + | as surface | ||
</source> | </source> | ||
[[Category:Performance]] | [[Category:Performance]] | ||
Latest revision as of 13:55, 16 March 2018
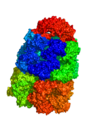
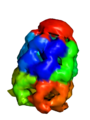
PyMOL can render very large (huge) surfaces of proteins, using a few tricks. First off, you should know that because of the size, you do not get the level of accuracy that you would for a smaller molecule. However, if you need to represent a molecule (or object) with a few million atoms, this may be your option of choice.
Example
The output from the example below is shown below. The PDB 1AON has nearly 60,000 atoms. This isn't the largest PDB, but it's a good example.
Strategy 1: Low resolution Gaussian Map
# load a whopping big PDB
fetch 1aon, struct, async=0
# create a color spectrum over the object
spectrum count, selection=struct
# === now create a pseudo-fcalc map (a 3D volumetric scalar field) ===
# set the B-factors nice and high for smoothness
set gaussian_b_floor, 40
# ~10 A map resolution
set gaussian_resolution, 10
# ~10 A map spacing with a 10 A surrounding buffer
# (you may need to vary this)
map_new map, gaussian, 10, struct, 10
# create a surface from the map
isosurface surf, map, 1.0
# now color the map based on the underlying protein
ramp_new ramp, struct, [0,10], [atomic, atomic]
color ramp, surf
# hide the ramp
disable ramp
Strategy 2: CA-only model with oversized radii
# load a whopping big PDB
fetch 1aon, struct, async=0
# reduce to CA atoms only
remove struct & not guide
# create a color spectrum over the object
spectrum count, selection=struct
# increase VDW radii to compensate volume of missing non-CA atoms
alter struct, vdw=4
# increase VDW radius of solvent
set solvent_radius, 4, struct
# show molecular surface
as surface