Difference between revisions of "Group"
Jump to navigation
Jump to search
| Line 1: | Line 1: | ||
| − | |||
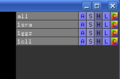
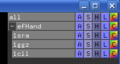
The [[Group]] command creates or updates a "group" object. The grouped objects are collected underneath a '''+''' sign in the object tree (see images). | The [[Group]] command creates or updates a "group" object. The grouped objects are collected underneath a '''+''' sign in the object tree (see images). | ||
| Line 19: | Line 18: | ||
</source> | </source> | ||
| − | == | + | == Examples == |
| + | |||
| + | === Creating, opening and closing === | ||
<source lang="python"> | <source lang="python"> | ||
group efHand, 1cll 1ggz 1sra | group efHand, 1cll 1ggz 1sra | ||
| Line 29: | Line 30: | ||
</source> | </source> | ||
| + | === More advanced usage of groups and naming === | ||
| + | |||
| + | <source lang="python"> | ||
| + | # names with dots are treated special | ||
| + | |||
| + | set group_auto_mode, 2 | ||
| + | |||
| + | # load the example protein | ||
| + | |||
| + | load $TUT/1hpv.pdb, 1hpv.other | ||
| + | |||
| + | # create the new entry called ".protein" in group 1hpv | ||
| + | |||
| + | extract 1hpv.protein, 1hpv.other and polymer | ||
| + | |||
| + | # create ".ligand in the 1hpv group | ||
| + | |||
| + | extract 1hpv.ligand, 1hpv.other and organic | ||
| + | |||
| + | # supports wildcards | ||
| + | |||
| + | show sticks, *.ligand | ||
| + | |||
| + | hide lines, *.protein | ||
| + | |||
| + | show surface, *.protein within 6 of *.ligand | ||
| + | |||
| + | show lines, byres *.protein within 4 of *.ligand | ||
| + | |||
| + | set two_sided_lighting | ||
| + | |||
| + | set transparency, 0.5 | ||
| + | |||
| + | set surface_color, white | ||
| + | |||
| + | # Also, to lexicographically sort the names in the control panel: | ||
| + | |||
| + | order *, yes | ||
| + | </source> | ||
== Notes == | == Notes == | ||
Group objects can usually be used as arguments to commands. It can be processed as a group or as a selection, in which case all the atoms from all objects in the group will be used. | Group objects can usually be used as arguments to commands. It can be processed as a group or as a selection, in which case all the atoms from all objects in the group will be used. | ||
Revision as of 10:36, 17 February 2011
The Group command creates or updates a "group" object. The grouped objects are collected underneath a + sign in the object tree (see images).
Group is tremendously helpful with multi-state or multi-structure sessions. Wildcards work great, for example:
# put all of objState into the group "ensemble".
group ensemble, objState*
Usage
group name, members, action
Examples
Creating, opening and closing
group efHand, 1cll 1ggz 1sra
# allow addition and removal from the group
group efHand, open
# disallow addition/removal from the group
group efHand, close
More advanced usage of groups and naming
# names with dots are treated special
set group_auto_mode, 2
# load the example protein
load $TUT/1hpv.pdb, 1hpv.other
# create the new entry called ".protein" in group 1hpv
extract 1hpv.protein, 1hpv.other and polymer
# create ".ligand in the 1hpv group
extract 1hpv.ligand, 1hpv.other and organic
# supports wildcards
show sticks, *.ligand
hide lines, *.protein
show surface, *.protein within 6 of *.ligand
show lines, byres *.protein within 4 of *.ligand
set two_sided_lighting
set transparency, 0.5
set surface_color, white
# Also, to lexicographically sort the names in the control panel:
order *, yes
Notes
Group objects can usually be used as arguments to commands. It can be processed as a group or as a selection, in which case all the atoms from all objects in the group will be used.