FocalBlur
Description
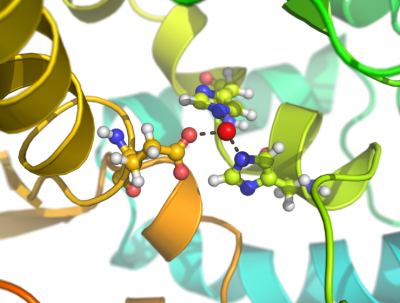
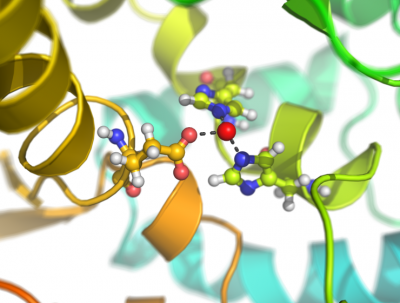
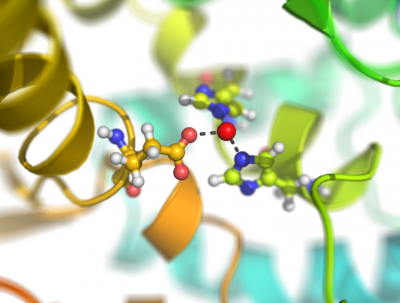
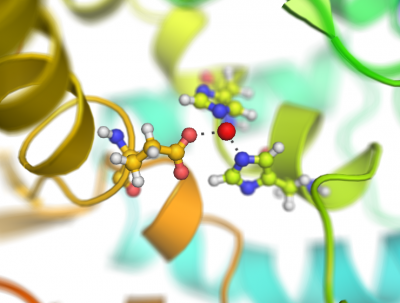
This script creates fancy figures by introducing a focal blur to the image. The object at the origin will be in focus.
Usage
load the script using the run command
FocalBlur(aperture=2.0,samples=100,raytrace=True)
For additional options, see the script comments.
Notes
- When using raytracing, the image creation will take n times longer than normal, where n is the number of samples.
- The script uses ImageMagick for creating the blured image. It has only been tested on Linux
- The aperture is a purely arbitrary value and not related to f stops on a camera.
- There is a bug preventing custom size images when not using raytracing.
Examples
Script
load the script using the run command
import random
from pymol import cmd
from os import system
from tempfile import mkdtemp
from shutil import rmtree
from math import sin,cos,pi
print 'Usage: FocalBlur(aperture=float,samples=int,raytrace=True/False,width=int,height=int)'
def FocalBlur(aperture=2.0,samples=10,raytrace=False,width=0,height=0):
'''
AUTHOR
Jarl Underhaug
University of Bergen
jarl_dot_underhaug_at_gmail_dot_com
USAGE
FocalBlur(aperture=float,samples=int,raytrace=True/False,width=int,height=int)
EXAMPELS
FocalBlur(aperture=1,samples=100,raytrace=False)
FocalBlur(aperture=2,samples=100,raytrace=True,width=600,height=400)
'''
# Because of a bug, only custom sizes when raytracing
if not raytrace:
width=0
height=0
# Create a temporary directory
tmpdir = mkdtemp()
# Get the orientation of the protein and the light
light = cmd.get('light')[1:-1]
light = [float(s) for s in light.split(',')]
view = cmd.get_view()
# Rotate the protein and the light in order to create the blur
for frame in range(samples):
# Angles to rotate protein and light
x = (random.random()-0.5)*aperture
y = (random.random()-0.5)*aperture
xr = x/180.0*pi
yr = y/180.0*pi
# Rotate the protein
cmd.turn('x',x)
cmd.turn('y',y)
# Rotate the light
ly = light[1]*cos(xr)-light[2]*sin(xr)
lz = light[2]*cos(xr)+light[1]*sin(xr)
lx = light[0]*cos(yr)+lz*sin(yr)
lz = lz*cos(yr)-lx*sin(yr)
cmd.set('light',[lx,ly,lz])
# Save the image
cmd.png(tmpdir+'/frame-%04d.png' % (frame),width=width,height=height,ray=raytrace)
# Return the protein and the light to the original orientation
cmd.set('light',light)
cmd.set_view(view)
# Create a blured image of all the frames
system('convert %s/frame-*.png +matte -average %s/blur.png' % (tmpdir,tmpdir))
# Load the blured image
cmd.load('%s/blur.png' % (tmpdir))
# Delete the temporary files
rmtree(tmpdir)