Difference between revisions of "Elbow angle"
Jaredsampson (talk | contribs) (→Examples: added more details to first example, and better explanation for Pymol-script-repo example) |
|||
| Line 10: | Line 10: | ||
== Examples == | == Examples == | ||
| + | Basic usage: | ||
<syntaxhighlight lang="python"> | <syntaxhighlight lang="python"> | ||
| − | # load an antibody Fab from the PDB | + | # load an antibody Fab fragment from the PDB |
fetch 3ghe, async=0 | fetch 3ghe, async=0 | ||
# calculate the elbow angle and draw the vectors | # calculate the elbow angle and draw the vectors | ||
elbow_angle 3ghe, draw=1 | elbow_angle 3ghe, draw=1 | ||
</syntaxhighlight> | </syntaxhighlight> | ||
| + | Note that if you don't specify the light/heavy chains or the variable domain limits, the default (L/H and 107/113 respectively) will be assumed. If your antibody is not Kabat or Chothia numbered, this will result in an incorrect value! Have a look at Dr. Andrew Martin's [http://www.bioinf.org.uk/abs/abnum/ ABNUM] for more information. | ||
| − | + | ||
| + | There is also an example .pml file in the Pymol-script-repo/examples directory which can be run by the following: | ||
<syntaxhighlight lang="python"> | <syntaxhighlight lang="python"> | ||
import ex | import ex | ||
Revision as of 17:22, 14 June 2012
| Type | Python Script |
|---|---|
| Download | elbow_angle.py |
| Author(s) | Jared Sampson |
| License | GPLv3 |
| This code has been put under version control in the project Pymol-script-repo | |
Introduction
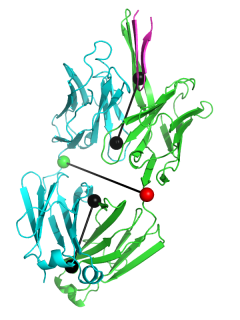
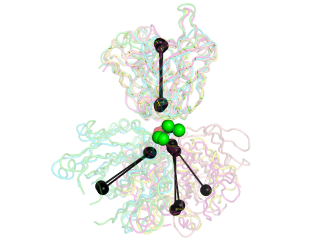
This script allows you to calculate the elbow angle of an antibody Fab fragment object and optionally draw a graphical representation of the vectors used to calculate the elbow angle.
Examples
Basic usage:
# load an antibody Fab fragment from the PDB
fetch 3ghe, async=0
# calculate the elbow angle and draw the vectors
elbow_angle 3ghe, draw=1
Note that if you don't specify the light/heavy chains or the variable domain limits, the default (L/H and 107/113 respectively) will be assumed. If your antibody is not Kabat or Chothia numbered, this will result in an incorrect value! Have a look at Dr. Andrew Martin's ABNUM for more information.
There is also an example .pml file in the Pymol-script-repo/examples directory which can be run by the following:
import ex
ex elbow_angle
The black "dumbbells" pass through the centers of mass of the variable and constant domains of each Fab. The green and red dumbbells denote the residues used to split the variable and constant domains, with a green ball for the light chain, and a red ball for the heavy chain.