Difference between revisions of "DistancesRH"
m |
m (download link and license update) |
||
| (6 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{Infobox script-repo | {{Infobox script-repo | ||
|type = script | |type = script | ||
| − | |download = | + | |download = [http://dx.doi.org/10.6084/m9.figshare.1031580 figshare] |
|author = [[User:PietroGattiLafranconi|Pietro Gatti-Lafranconi]] | |author = [[User:PietroGattiLafranconi|Pietro Gatti-Lafranconi]] | ||
| − | |license = [http://creativecommons.org/licenses/by | + | |license = [http://creativecommons.org/licenses/by/4.0/ CC BY 4.0] |
}} | }} | ||
| Line 16: | Line 16: | ||
* display distance objects in pymol | * display distance objects in pymol | ||
* restrict distances between hydrogen-bond forming atoms only | * restrict distances between hydrogen-bond forming atoms only | ||
| + | |||
The script also runs a number of controls to verify that inputs and outputs exists/are suitable. | The script also runs a number of controls to verify that inputs and outputs exists/are suitable. | ||
| Line 21: | Line 22: | ||
== Usage == | == Usage == | ||
| − | |||
distancesRH obj, ref, dist, [chain, [output S/P/N, [show Y/N, [Hbonds Y/N, [aname]]]]] | distancesRH obj, ref, dist, [chain, [output S/P/N, [show Y/N, [Hbonds Y/N, [aname]]]]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
== Required Arguments == | == Required Arguments == | ||
| − | |||
* '''obj''' = object name | * '''obj''' = object name | ||
* '''ref''' = reference amino acid id | * '''ref''' = reference amino acid id | ||
| Line 45: | Line 40: | ||
== Examples == | == Examples == | ||
| − | example #1 | + | '''example #1''' |
<source lang="python"> | <source lang="python"> | ||
| − | distancesRH 2WHH, 25, 3.2 | + | distancesRH 2WHH, 25, 3.2, show=Y |
</source> | </source> | ||
| − | Will show distances between all atoms of residue 25 in 2WHH and any atom within 3.2 Å and return the result on screen. | + | Will show distances between all atoms of residue 25 in 2WHH and any atom within 3.2 Å and return the result on screen (figure 1, left). |
| + | <source lang="python"> | ||
| + | distancesRH 2WHH, 25, 3.2, show=Y, aname=OD1, output=P | ||
| + | </source> | ||
| + | Same result as before, but only distances from atom OD1 are measured and shown. Results will be printed in 'distancesRH_25-OD1.txt' (figure 1, right). | ||
| + | [[File:distancesRH_ex1.png|450px|example #1]] | ||
| − | example #2 | + | |
| + | '''example #2''' | ||
| + | <source lang="python"> | ||
| + | distancesRH 1EFA, 901, 3.5, show=Y | ||
| + | </source> | ||
| + | Will show distances between all atoms in resi 901 of 1EFA and all atoms within 3.5 Å (figure 2, left). | ||
<source lang="python"> | <source lang="python"> | ||
| − | distancesRH | + | distancesRH 1EFA, 901, 3.5, show=Y, Hbonds=Y |
</source> | </source> | ||
| − | + | Distances will now be calculated between atoms that can form H bonds only (figure 2, right). | |
| + | |||
| + | [[File:distancesRH_ex2.png|450px|example #2]] | ||
| + | |||
| + | |||
| + | '''example #3''' | ||
| + | <source lang="python"> | ||
| + | distancesRH 1EFA, 18, 4.7, chain=B, show=Y, aname=NE2 | ||
| + | </source> | ||
| + | Distances between atom NE2 of residue 19 in chain B of 1EFA and all atoms within 4.7 Å (all of which are in the DNA fragment of chain D, figure 3). | ||
| + | |||
| + | [[File:distancesRH_ex3.png|300px|example #3]] | ||
| + | |||
| + | |||
| + | == Notes, Further Developments and Requests == | ||
| + | * This script works with visible objects (handling of multiple states will hopefully be implemented soon) | ||
| + | * As it is intended to be user-friendly more than fast, this script is not suitable to calculate distances between large objects. Use [[Quick_dist]] instead. | ||
| + | |||
| + | |||
| + | == Updates == | ||
| + | * '''11/01/2013''' - edit #1: ''distances measured between different chains'' | ||
| + | * '''13/01/2013''' - edit #2: ''print output optimsed'' | ||
== The Code == | == The Code == | ||
| + | |||
| + | Copy the following text and save it as distancesRH.py | ||
| + | |||
<source lang="python"> | <source lang="python"> | ||
| + | |||
| + | """ | ||
| + | DESCRIPTION | ||
| + | Given an object, a reference amino acid and a radius, this scripts calculates pairwise | ||
| + | distances between all atoms in the reference and all atoms that fall within the given range. | ||
| + | Distances are either returned on screen (default) or printed on file. | ||
| + | Distances can be calculated from a single atom only. | ||
| + | Optionally, only atoms that can form H-bonds can be returned (but check distances!) | ||
| + | |||
| + | More information at: PymolWiki | ||
| + | http://pymolwiki.org/index.php/DistancesRH | ||
| + | |||
| + | AUTHOR | ||
| + | Pietro Gatti-Lafranconi, 2012 | ||
| + | Please inform me if you use/improve/like/dislike/publish with this script. | ||
| + | CC BY-NC-SA | ||
| + | """ | ||
from pymol import cmd, stored, math | from pymol import cmd, stored, math | ||
| Line 93: | Line 139: | ||
cmd.select("RefAtom", cen+refA) | cmd.select("RefAtom", cen+refA) | ||
m1=cmd.get_model ("RefAtom") | m1=cmd.get_model ("RefAtom") | ||
| − | cmd.select("Atoms", cen+refA+" around "+str(dist)+" and not resi "+ref+" and "+ | + | cmd.select("Atoms", cen+refA+" around "+str(dist)+" and not resi "+ref+" and "+obj) |
cmd.show("nb_spheres", "(Atoms or Reference) and (resn HOH or HET)") | cmd.show("nb_spheres", "(Atoms or Reference) and (resn HOH or HET)") | ||
cmd.select("Residues", "byres Atoms") | cmd.select("Residues", "byres Atoms") | ||
| Line 155: | Line 201: | ||
print screenOP | print screenOP | ||
if output=="P": | if output=="P": | ||
| + | if aname=="*": aname="ALL" | ||
print 'data printed in "distances_rH_'+ref+'-'+aname+'.txt" file' | print 'data printed in "distances_rH_'+ref+'-'+aname+'.txt" file' | ||
f=open('distances_rH_'+ref+'-'+aname+'.txt','w') | f=open('distances_rH_'+ref+'-'+aname+'.txt','w') | ||
Latest revision as of 12:56, 22 May 2014
| Type | Python Script |
|---|---|
| Download | figshare |
| Author(s) | Pietro Gatti-Lafranconi |
| License | CC BY 4.0 |
Introduction
Given an object, a reference amino acid and a radius, this scripts calculates pairwise distances between atom(s) in the reference and all atoms that fall within the given range.
The script is intended to provide a range of useful tools while requiring minimal coding skills (i.e. it is not optimised for efficiency).
By changing input parameters it is possible to:
- choose the output (none, on screen, on file)
- measure distances from one single atom
- display distance objects in pymol
- restrict distances between hydrogen-bond forming atoms only
The script also runs a number of controls to verify that inputs and outputs exists/are suitable.
Usage
distancesRH obj, ref, dist, [chain, [output S/P/N, [show Y/N, [Hbonds Y/N, [aname]]]]]
Required Arguments
- obj = object name
- ref = reference amino acid id
- dist = radius in Angstroms
Optional Arguments
- chain = selected chain
- output = accepts S (screen), P (print) or N (none), default=S
- show = shows distances in pymol window, default=N
- Hbonds = restricts results to atoms that can form Hbonds, default=N
- aname = selects a specific atom in reference object and to calculate distances from one individual atom
Examples
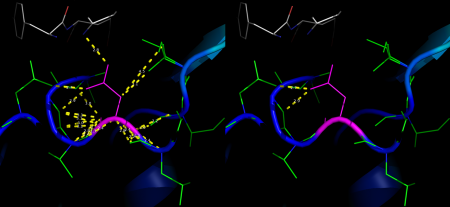
example #1
distancesRH 2WHH, 25, 3.2, show=Y
Will show distances between all atoms of residue 25 in 2WHH and any atom within 3.2 Å and return the result on screen (figure 1, left).
distancesRH 2WHH, 25, 3.2, show=Y, aname=OD1, output=P
Same result as before, but only distances from atom OD1 are measured and shown. Results will be printed in 'distancesRH_25-OD1.txt' (figure 1, right).
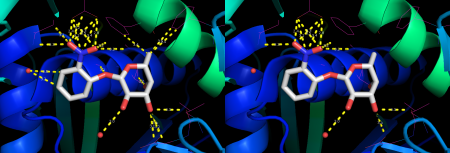
example #2
distancesRH 1EFA, 901, 3.5, show=Y
Will show distances between all atoms in resi 901 of 1EFA and all atoms within 3.5 Å (figure 2, left).
distancesRH 1EFA, 901, 3.5, show=Y, Hbonds=Y
Distances will now be calculated between atoms that can form H bonds only (figure 2, right).
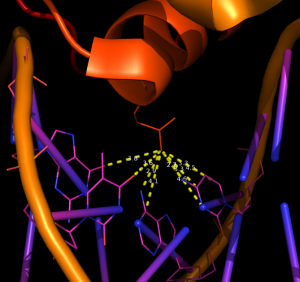
example #3
distancesRH 1EFA, 18, 4.7, chain=B, show=Y, aname=NE2
Distances between atom NE2 of residue 19 in chain B of 1EFA and all atoms within 4.7 Å (all of which are in the DNA fragment of chain D, figure 3).
Notes, Further Developments and Requests
- This script works with visible objects (handling of multiple states will hopefully be implemented soon)
- As it is intended to be user-friendly more than fast, this script is not suitable to calculate distances between large objects. Use Quick_dist instead.
Updates
- 11/01/2013 - edit #1: distances measured between different chains
- 13/01/2013 - edit #2: print output optimsed
The Code
Copy the following text and save it as distancesRH.py
"""
DESCRIPTION
Given an object, a reference amino acid and a radius, this scripts calculates pairwise
distances between all atoms in the reference and all atoms that fall within the given range.
Distances are either returned on screen (default) or printed on file.
Distances can be calculated from a single atom only.
Optionally, only atoms that can form H-bonds can be returned (but check distances!)
More information at: PymolWiki
http://pymolwiki.org/index.php/DistancesRH
AUTHOR
Pietro Gatti-Lafranconi, 2012
Please inform me if you use/improve/like/dislike/publish with this script.
CC BY-NC-SA
"""
from pymol import cmd, stored, math
def distancesRH (obj, ref, dist, chain=0, output="S", show="N", Hbonds="N", aname="*"):
"""
usage: distancesRH obj, ref, dist, [chain, [output S/P/N, [show Y/N, [Hbonds Y/N, [aname]]]]]
obj: object name, ref:reference aa, dist: radius in A, chain: selected chain
output: accepts S (screen), P (print) or N (none), default=S
show: shows distances in pymol window, default=N
Hbonds: restricts results to atoms that can form Hbonds, default=N
aname: name of a specific atom in ref (to calculate distances from one atom only)
example: distancesRH 2WHH, 25, 3.2, [B, [S, [N, [Y, [CB]]]]]
"""
print ""
#delete previous selections and displayed distances
cmd.delete ("dist*")
cmd.delete ("Atoms")
cmd.delete ("Residues")
cmd.delete ("Reference")
cmd.delete ("Hbonds")
cmd.delete ("RefAtom")
#creates and names selections
if chain==0: cen=obj
else: cen=obj+" and chain "+chain
refA=" and resi "+ref+" and name "+aname
cmd.select("Reference", "byres "+cen+refA)
m1=cmd.get_model ("Reference")
if aname!="*":
cmd.select("RefAtom", cen+refA)
m1=cmd.get_model ("RefAtom")
cmd.select("Atoms", cen+refA+" around "+str(dist)+" and not resi "+ref+" and "+obj)
cmd.show("nb_spheres", "(Atoms or Reference) and (resn HOH or HET)")
cmd.select("Residues", "byres Atoms")
m2=cmd.get_model("Atoms and visible")
if Hbonds=="Y":
cmd.select("__Hbonds1", "Reference and not symbol c")
if aname!="*":
cmd.select("__Hbonds1", "RefAtom and not symbol c")
cmd.select("__Hbonds2", "Atoms and not symbol c")
cmd.select("Hbonds", "__Hbonds1 or __Hbonds2")
cmd.show("lines", "Hbonds")
m1=cmd.get_model ("__Hbonds1")
m2=cmd.get_model ("__Hbonds2 and visible")
#controllers
if (not (output=="S" or output=="N" or output=="P")) or (not (show=="Y" or show=="N")) or (not (Hbonds=="Y" or Hbonds=="N")):
print "Malformed input, please try again"
return
abort=1
abort2=0
mc=cmd.get_model(cen)
for c0 in range(len(mc.atom)):
if mc.atom[c0].resi==ref: abort=0
mc2=cmd.get_model(cen+refA)
if aname!="*":
abort2=1
for c00 in range (len(mc2.atom)):
if mc2.atom[c00].name==aname: abort2=0
if Hbonds=="Y":
if aname[0]=="C":
print "Warning: no atoms in the selection can form H-bonds"
return
if abort==1: print "Warning: no such residue in the reference object"
if abort2==1:
if abort==0: print "Warning: no such atom in the reference object"
if abort==1 or abort2==1: return
#measures distances
distance2=[]
distances=[]
screenOP=""
fileOP=""
for c1 in range(len(m1.atom)):
for c2 in range(len(m2.atom)):
if math.sqrt(sum(map(lambda f: (f[0]-f[1])**2, zip(m1.atom[c1].coord,m2.atom[c2].coord))))<float(dist):
distance=cmd.distance (obj+"//"+m1.atom[c1].chain+"/"+m1.atom[c1].resi+"/"+m1.atom[c1].name, obj+"//"+m2.atom[c2].chain+"/"+m2.atom[c2].resi+"/"+m2.atom[c2].name)
distances.append(distance)
screenOP+="%s/%s/%s/%s to %s/%s/%s/%s: %.3f\n" % (m1.atom[c1].chain,m1.atom[c1].resn,m1.atom[c1].resi,m1.atom[c1].name,m2.atom[c2].chain,m2.atom[c2].resn,m2.atom[c2].resi,m2.atom[c2].name, distance)
fileOP+="%s/%s/%s/%s\t\t\t%s/%s/%s/%s\t\t\t\t%.5f\n"% (m1.atom[c1].chain,m1.atom[c1].resn,m1.atom[c1].resi,m1.atom[c1].name,m2.atom[c2].chain,m2.atom[c2].resn,m2.atom[c2].resi,m2.atom[c2].name, distance)
#last controller
if len(distances)==0:
if abort2==0:
print "Warning: no atoms within the selected range"
return
#data outputs
if output=="N": print "Data recorded, but not printed"
if output=="S":
print "Distance of atoms in "+obj+"/"+ref+"/"+aname+" to all atoms within "+dist+" Angstroms"
if Hbonds=="Y": print "Distances restricted to potential H-bond pairs"
print screenOP
if output=="P":
if aname=="*": aname="ALL"
print 'data printed in "distances_rH_'+ref+'-'+aname+'.txt" file'
f=open('distances_rH_'+ref+'-'+aname+'.txt','w')
f.write("Pairwise distances betweet atoms in %s and all atoms within %s Angstroms\n" % (obj+"/"+ref+"/"+aname, dist))
if Hbonds=="Y": f.write("Distances restricted to potential H-bond pairs\n")
f.write("Atom in reference\tAtom in destination\t\tdistance\n")
f.write(fileOP)
f.close()
print "All done! Good luck with your data"
#graphical output
if show=="N": cmd.delete ("dist*")
if show=="Y":
cmd.show("lines", "Reference")
cmd.show("lines", "Residues")
cmd.extend("distancesRH", distancesRH);