Difference between revisions of "Displacementmap"
m (moved DisplacementMap to Displacementmap: Small letters) |
|||
| Line 30: | Line 30: | ||
== Example == | == Example == | ||
| − | < | + | <syntaxhighlight lang="python"> |
dispmap(molecule1="NIL", molecule2="NIL", mindist=30.0, mindelta=15.0, resi1=str(0), resi2=str(0), atom='CA', listlength=5, showsticks='yes'): | dispmap(molecule1="NIL", molecule2="NIL", mindist=30.0, mindelta=15.0, resi1=str(0), resi2=str(0), atom='CA', listlength=5, showsticks='yes'): | ||
dispmap Open5NT, Closed5NT, 40.0, 15.0, resi1=206, resi2=1-512.515-550 | dispmap Open5NT, Closed5NT, 40.0, 15.0, resi1=206, resi2=1-512.515-550 | ||
dispmap Open5NT, Closed5NT, 30.0, 15.0, resi2=1-512.515-550, atom=CA, listlength=10 | dispmap Open5NT, Closed5NT, 30.0, 15.0, resi2=1-512.515-550, atom=CA, listlength=10 | ||
| − | </ | + | </syntaxhighlight> |
== Output == | == Output == | ||
Revision as of 11:33, 1 December 2011
Author
This pymol script is made by Troels Emtekær Linnet
Overview
DisplacementMap is made for easy investigations of suitable positions for site-directed mutagenesis of amino residues into cysteines and FRET/EPR pair labelling.
A Open and Closed form of a protein should be loaded. New objects should be created for the selected asymmetric unit. Parts of the protein should be aligned, leaving the flexible part in two different positions.
The input is the objects, Open (molecule1) and Closed (molecule2).
Further is the criteria for selecting which atom the distance should be calculated between. Standard is atom='CA' (atom).
Then one selects the Förster distance R0 (mindist). This is the minimum distance between the residues. This depends of the selection of the FRET pair and protein at hand. But usually in the range 40 - 80 Angstrom is suitable.
Then one defines the minimum displacement that is accepted. Usually R0/2 (mindelta).
The script will find the 5 best (listlength=5) positive and negative distance displacement between the two objects.
It parses the results back to Pymol, that is standard set to show it as sticks (showsticks='yes').
If one is looking for a particular residue range for the FRET pair, this can be specified with two input.
resi1=24.45-47.86 resi2=100-105.107 resi1 is "from" and resi2 is "to". Individual residues are split by a ".", and ranges are defined with "-".
In the end, it makes a large data-matrix with all the distances. It also produces a gnuplot file, for easy visualisation. Just drag the .plt file for win gnuplot command window and it plots your datamatrix.
Bugs
If the criterion is set to low, the memory gets flooded in the data-matrix file, making the file unreadable. No solutions found yet.
Instructions
- Make a folder
- Copy the code to your machine, and name: dispmap.py . You can for example put it into your pymol search path.
- Download the .pdb files of the Open and Closed form of your protein
- Make a pymol script file, that makes the alignment and such. See example.
- Run the script and see the results in command window and suggestions in pymol window
- Run the gnuplot file to see the data-matrix
Example
dispmap(molecule1="NIL", molecule2="NIL", mindist=30.0, mindelta=15.0, resi1=str(0), resi2=str(0), atom='CA', listlength=5, showsticks='yes'):
dispmap Open5NT, Closed5NT, 40.0, 15.0, resi1=206, resi2=1-512.515-550
dispmap Open5NT, Closed5NT, 30.0, 15.0, resi2=1-512.515-550, atom=CA, listlength=10
Output
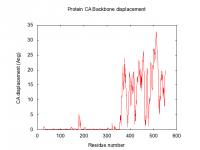
Suggestions are created in pymol, and gnuplot file is created for easy visualisation of pair data-matrix and the general backbone displacement.
Text output
In the data-matrix.txt file, you find the best suggestions
# Input 1: Open5NT and Input 2: Closed5NT
# Find for: CA with min. residue-residue dist: 30.0 Angstrom
# Looking for min. displacement dist: 15.0 Angstrom
# I give nr# suggestions: 5, and do I show sticks in pymol?: yes
# I look for suggestions in the range: ([0]=>means all residues)
# for Input 1: ['0'] and for Input 2: ['0']
# Mutation info is BLOSUM62 log-odds likelihood score and PAM250 is probability in % for evolutionary distance
###########################################################################################################
# Max Negative and positive distances # Mutation info #
###########################################################################################################
# Obj.1 Obj.2 Delta Op-Op Cl-Cl # Obj.1 Obj.2 Delta Op-Op Cl-Cl # Res.1 Res.2 # Res.1 Res.2 #
# Res.1 Res.2 -Dist Dist Dist # Res.1 Res.2 +Dist Dist Dist # B62/PAM250% # B62/PAM250% #
###########################################################################################################
# PRO241 ASP456 -25.7 59.1 33.4 # PRO274 PRO513 26.8 31.2 58.0 # -3/ 2 -3/ 1 # -3/ 2 -3/ 2 #
# LYS197 ASP456 -25.6 57.3 31.7 # THR273 PRO513 26.1 31.6 57.7 # -1/ 1 -3/ 1 # -1/ 2 -3/ 2 #
# PRO513 ASP456 -25.4 32.4 7.0 # PRO274 GLY514 24.8 32.9 57.6 # -3/ 2 -3/ 1 # -3/ 2 -3/ 2 #
# LEU198 ASP456 -25.3 59.0 33.7 # PRO274 LYS512 24.7 30.3 55.0 # -1/ 1 -3/ 1 # -3/ 2 -1/ 1 #
# GLN201 ASP456 -25.2 62.8 37.6 # ASN311 PRO513 24.7 35.6 60.3 # -3/ 1 -3/ 1 # -3/ 1 -3/ 2 #
The script also automatically make the gnuplot plot file (.plt), with all the defined variables, for easy visualisation of the data-matrix.txt and the backbone displacement.
reset
cd '/homes/linnet/Documents/Speciale/5NT-project/Mutant-construct/Distance-Plot/dispmap'
#Title hacks \n is newline, and 0,1 is x,y offset adjustment
set title "Protein CA Displacement matrix map \n ResRes min. 30.0 Ang, Delta min. 15.0 Ang" 0,1
# x is column
set xlabel 'Res nr. for Closed5NT'
# y is row
set ylabel 'Res nr. for Open5NT'
#set xrange [300:550]; set yrange [0:400]
#set xtics 50
#set ytics 50
#set mxtics 5
#set mytics 5
set size ratio 1
unset key
set cbrange [-30:30]
set palette defined (-30 'blue', 0 'white', 30 'red')
set pm3d map
#set term postscript eps enhanced color
#set output "Open5NT-Closed5NT-CA-dist.eps"
set term png
set output "Open5NT-Closed5NT-CA-dist.png"
splot 'Open5NT-Closed5NT-CA-dist.txt' matrix
#For the backbone displacement
set title "Protein CA Backbone displacement" 0,1
set xlabel 'Residue number'
set ylabel 'CA displacement (Ang)'
#set xrange [0:550]; set yrange [0:40]
#set xtics 50
#set ytics 10
#set mxtics 5
#set mytics 5
set size ratio 0.75
unset key
#set term postscript eps enhanced color
#set output "Open5NT-Closed5NT-CA-dist-backbone.eps"
set term png
set output "Open5NT-Closed5NT-CA-dist-backbone.png"
plot 'Open5NT-Closed5NT-CA-dist-backbone.txt' using 1:2 title 'Backbone displacement' with lines
Pymol script file
cd /homes/linnet/Documents/Speciale/5NT-project/Mutant-construct/Distance-Plot/dispmap
#C:\Users\tlinnet\Documents\My Dropbox\Speciale\5NT-project\Mutant-construct\Distance-Plot\dispmap
### load pdb files and rename
fetch 1HP1, async=0
fetch 1HPU, async=0
hide everything, all
### Select asymmetric units from pdb file
create Open5NT, /1HP1//A
create Closed5NT, /1HPU//A
delete 1HP1
delete 1HPU
cartoon auto
show cartoon, all
set cartoon_fancy_helices=1
set bg,[1,1,1]
### align
#align Open5NT and resi 26-355, Closed5NT and resi 26-355
# Super is must faster than align http://www.pymolwiki.org/index.php/Super
super Open5NT and resi 26-355, Closed5NT and resi 26-355
set auto_zoom, off
set_view (\
-0.374262989, 0.692084968, -0.617209554,\
-0.681849480, -0.656483948, -0.322660774,\
-0.628496349, 0.300085038, 0.717594206,\
0.000000000, 0.000000000, -258.556884766,\
-0.613845825, 0.472507477, 1.410455704,\
205.729583740, 311.384277344, 0.000000000 )
### Color
set_color goldenrod1, [1.000, 0.757, 0.145]
color goldenrod1, resi 26-355
set_color darkolivegreen1, [0.792, 1.000, 0.439]
color darkolivegreen1, Open5NT and resi 356-362
set_color darkolivegreen4, [0.431, 0.545, 0.239]
color darkolivegreen4, Closed5NT and resi 356-362
set_color chocolate3, [0.804, 0.400, 0.114]
color chocolate3, Open5NT and resi 363-550
set_color purple4, [0.333, 0.102, 0.545]
color purple4, Closed5NT and resi 363-550
# Select active site
select active_site, resi 117+120+252+116+217+84+41+43+254
show sticks, active_site
# Make Cys-Cys bonds
create SS, (cys/ca+cb+sg) and byres (cys/sg and bound_to cys/sg)
show sticks, SS
color yellow, SS
# Mark water molecules
create waters, resn HOH
show nb_spheres, waters
color blue, waters
disable waters
### Make sharper, and transparent
set fog=0
set cartoon_transparency, 0.7
### Load the function
import dispmap
dispmap
#dispmap Open5NT, Closed5NT, 40.0, 15.0, resi1=206, resi2=1-512.515-550
## Look for serine
#dispmap mindist=40.0, mindelta=15.0, resi1=206, resi2=330.347.350.405.412.419.457.467.533.534.539.548.336.367.383.397.439.448.490.495.501.518
#dispmap resi1=308, resi2=513
Python Code
The code can be downloaded fast from here http://tinyurl.com/pymoldispmap
- wget http://tinyurl.com/pymoldispmap
- mv pymoldispmap dispmap.py
from pymol import cmd, stored, selector
from math import *
import os, re
## Thx for inspiration from Andreas Henschel
## http://www.mail-archive.com/pymol-users@lists.sourceforge.net/msg05595.html (17 dec 2010)
## And from Simple scriptin PymMOl http://www.pymolwiki.org/index.php/Simple_Scripting
### Ma.Sc student. Troels Linnet, 2010-12-18. troels.linnet@bbz.uni-leipzig.de
### Calculates the distance for example between all CA atoms between a closed and open form of a structure.
### Give a data matrix and a gnuplot file, and input to pymol for easy visualisation
### Possible so select interesting residues in ranges. Needs to be separated with a dot '.'
### Example input from pymol. with 2 objects.
### dispmap O5NT-1HP1-A, C5NT-1HPU-C, 30.0, 15.0, resi1=23-25, atom=CA, showsticks=yes
def dispmap(molecule1="NIL", molecule2="NIL", mindist=30.0, mindelta=15.0, resi1=str(0), resi2=str(0), atom='CA', listlength=5, showsticks='yes'):
if molecule1=="NIL":
assert len(cmd.get_names())!=0, "Did you forget to load a molecule? There are no objects in pymol."
molecule1=cmd.get_names()[0]
if molecule2=="NIL":
assert len(cmd.get_names())!=0, "Did you forget to load a molecule? There are no objects in pymol."
molecule2=cmd.get_names()[1]
print "You passed in %s and %s" % (molecule1, molecule2)
### Open filenames
filename = str(molecule1) + "-" + str(molecule2) + "-" + str(atom) + "-dist"
backbonefilename = str(molecule1) + "-" + str(molecule2) + "-" + str(atom) + "backbone-dist.txt"
outfile = open(filename+".txt", "w")
backboneoutfile = open(filename+"-backbone.txt", "w")
gnuoutfile = open(filename+".plt", "w")
print("I have opened matrix %s for you\n" % (filename+".txt"))
### Sorting for interesting residues for Obj1 and Obj2.
### Input is a string, and need to be sorted.
resi1 = resi1.split('.')
resi2 = resi2.split('.')
resi1List = []
resi2List = []
for i in resi1:
if '-' in i:
tmp = i.split('-')
resi1List.extend(range(int(tmp[0]),int(tmp[-1])+1))
if '-' not in i:
resi1List.append(int(i))
for i in resi2:
if '-' in i:
tmp = i.split('-')
resi2List.extend(range(int(tmp[0]),int(tmp[-1])+1))
if '-' not in i:
resi2List.append(int(i))
resi1List.sort()
resi2List.sort()
### Only take the lines where atom is specified in input
Object3 = molecule1 + " and name " + str(atom)
Object4 = molecule2 + " and name " + str(atom)
### Open 2 name lists
### Append residue and atom name to the lists
stored.OpenPDB = []
stored.ClosedPDB = []
cmd.iterate(Object3, "stored.OpenPDB.append((resi, name, resn))")
cmd.iterate(Object4, "stored.ClosedPDB.append((resi, name, resn))")
### Open 2 x,y,z position lists
### Append atom position
stored.OpenPos = []
stored.ClosedPos = []
cmd.iterate_state(1, selector.process(Object3), "stored.OpenPos.append((x,y,z))")
cmd.iterate_state(1, selector.process(Object4), "stored.ClosedPos.append((x,y,z))")
### Sometimes residues gets skipped in X-ray crys, because of low signal or sim. This leads to number conflict.
### Make ordered lists according to residue number. Find largest residue number via -1
OpenOrderedPDB = []
ClosedOrderedPDB = []
OpenOrderedPos = []
ClosedOrderedPos = []
BackboneDisp = []
### First fill lists with zeros
for i in range(int(stored.OpenPDB[-1][0])+1):
OpenOrderedPDB.append([0,0,0])
for i in range(int(stored.ClosedPDB[-1][0])+1):
ClosedOrderedPDB.append([0,0,0])
for i in range(int(stored.OpenPDB[-1][0])+1):
OpenOrderedPos.append((0,0,0))
for i in range(int(stored.ClosedPDB[-1][0])+1):
ClosedOrderedPos.append((0,0,0))
for i in range(int(stored.OpenPDB[-1][0])+1):
BackboneDisp.append([i,0,"NIL",atom])
### Fill in data the right places
j=0
for i in stored.OpenPDB:
OpenOrderedPDB[int(i[0])]=[int(i[0]),i[1],i[2]]
OpenOrderedPos[int(i[0])]=stored.OpenPos[j]
j = j + 1
j=0
for i in stored.ClosedPDB:
ClosedOrderedPDB[int(i[0])]=[int(i[0]),i[1],i[2]]
ClosedOrderedPos[int(i[0])]=stored.ClosedPos[j]
j = j + 1
### Make a list with the missing residues
MissingRes = []
for index, resi in enumerate(OpenOrderedPDB):
if abs(OpenOrderedPDB[index][0]-ClosedOrderedPDB[index][0]) != 0:
MissingRes.append(abs(OpenOrderedPDB[index][0]-ClosedOrderedPDB[index][0]))
print("Following residues miss in one of the files, and are discarded for")
print("further calculations")
print(MissingRes)
print("")
### Make the data matrix
CalcMatrix = create_nXn_matrix(len(OpenOrderedPos))
print("Calculating a %s X %s distance Matrix" % (len(OpenOrderedPos), len(ClosedOrderedPos)))
### Make a list with 10 most negative/positive distances
MaxNegDist = []
MaxPosDist = []
for i in range(int(listlength)):
MaxNegDist.append([0,0,0,0,0,0,0])
MaxPosDist.append([0,0,0,0,0,0,0])
### Calculate distances
for i in range(len(OpenOrderedPos)):
for j in range(len(ClosedOrderedPos)):
if OpenOrderedPos[i][0] != 0 and ClosedOrderedPos[j][0] != 0 and OpenOrderedPDB[i][0] not in MissingRes and ClosedOrderedPDB[j][0] not in MissingRes:
distOpenOpen = distance(OpenOrderedPos, OpenOrderedPos, i, j)
distClosedClosed = distance(ClosedOrderedPos, ClosedOrderedPos, i, j)
distOpenClosed = distance(OpenOrderedPos, ClosedOrderedPos, i, j)
DeltaDist = distOpenClosed - distOpenOpen
if i == j: BackboneDisp[i] = [i, DeltaDist, OpenOrderedPDB[i][2], atom]
###Test if distance is larger than threshold
if distOpenOpen >= float(mindist) and distClosedClosed >= float(mindist) and abs(DeltaDist) >= float(mindelta):
CalcMatrix[i][j] = str(round(DeltaDist, 0))
if DeltaDist < 0 and DeltaDist < MaxNegDist[-1][0] and (i in resi1List or resi1List[-1]==0) and (j in resi2List or resi2List[-1]==0):
MaxNegDist[-1][0] = DeltaDist
MaxNegDist[-1][1] = i
MaxNegDist[-1][2] = j
MaxNegDist[-1][3] = distOpenOpen
MaxNegDist[-1][4] = distOpenClosed
MaxNegDist[-1][5] = str(OpenOrderedPDB[i][2])
MaxNegDist[-1][6] = str(ClosedOrderedPDB[j][2])
MaxNegDist = sorted(MaxNegDist)
if DeltaDist > 0 and DeltaDist > MaxPosDist[-1][0] and (i in resi1List or resi1List[-1]==0) and (j in resi2List or resi2List[-1]==0):
MaxPosDist[-1][0] = DeltaDist
MaxPosDist[-1][1] = i
MaxPosDist[-1][2] = j
MaxPosDist[-1][3] = distOpenOpen
MaxPosDist[-1][4] = distOpenClosed
MaxPosDist[-1][5] = str(OpenOrderedPDB[i][2])
MaxPosDist[-1][6] = str(ClosedOrderedPDB[j][2])
MaxPosDist = sorted(MaxPosDist, reverse=True)
print("I made a datamatrix and backbone.txt file for you")
print("matrix: %s backbone: %s"%(filename+".txt",filename+"-backbone.txt"))
print("I made a gnuplot file for you, to view the datamatrix and the backbone displacement")
print("filename: %s\n"%(filename+".plt"))
###Print distance matrix
line01="# Input 1: %s and Input 2: %s"%(molecule1,molecule2)
line02="# Find for: %s with min. residue-residue dist: %s Angstrom"%(atom,mindist)
line03="# Looking for min. displacement dist: %s Angstrom"%(mindelta)
line04="# I give nr# suggestions: %s, and do I show sticks in pymol?: %s"%(listlength,showsticks)
line05="# I look for suggestions in the range: ([0]=>means all residues)"
line06="# for Input 1: %s and for Input 2: %s"%(resi1, resi2)
line07="# Mutation info is BLOSUM62 log-odds likelihood score and PAM250 is probability in % for evolutionary distance"
line08="###########################################################################################################"
line09="# Max Negative and positive distances # Mutation info #"
line10="# Obj.1 Obj.2 Delta Op-Op Cl-Cl # Obj.1 Obj.2 Delta Op-Op Cl-Cl # Res.1 Res.2 # Res.1 Res.2 #"
line11="# Res.1 Res.2 -Dist Dist Dist # Res.1 Res.2 +Dist Dist Dist # B62/PAM250% # B62/PAM250% #"
outfile.write(line01+'\n')
outfile.write(line02+'\n')
outfile.write(line03+'\n')
outfile.write(line04+'\n')
outfile.write(line05+'\n')
outfile.write(line06+'\n')
outfile.write(line07+'\n')
outfile.write(line08+'\n')
outfile.write(line09+'\n')
outfile.write(line08+'\n')
outfile.write(line10+'\n')
outfile.write(line11+'\n')
outfile.write(line08+'\n')
print(line01)
print(line02)
print(line03)
print(line04)
print(line05)
print(line06)
print(line07)
print(line08)
print(line09)
print(line08)
print(line10)
print(line11)
print(line08)
for i in range(len(MaxNegDist)):
text="# %3s%3s %3s%3s %5s %5s %5s # %3s%3s %3s%3s %5s %5s %5s # %2s/%2s %2s/%2s # %2s/%2s %2s/%2s #"%(MaxNegDist[i][5],MaxNegDist[i][1],MaxNegDist[i][6],MaxNegDist[i][2],round(MaxNegDist[i][0],1),round(MaxNegDist[i][3],1),round(MaxNegDist[i][4],1),MaxPosDist[i][5],MaxPosDist[i][1],MaxPosDist[i][6],MaxPosDist[i][2],round(MaxPosDist[i][0],1),round(MaxPosDist[i][3],1),round(MaxPosDist[i][4],1),cysb62(shortaa(str(MaxNegDist[i][5]))),pam250(shortaa(str(MaxNegDist[i][5]))),cysb62(shortaa(str(MaxNegDist[i][6]))),pam250(shortaa(str(MaxNegDist[i][6]))),cysb62(shortaa(str(MaxPosDist[i][5]))),pam250(shortaa(str(MaxPosDist[i][5]))),cysb62(shortaa(str(MaxPosDist[i][6]))),pam250(shortaa(str(MaxPosDist[i][6]))))
outfile.write(text+'\n')
print(text)
for i in range(len(CalcMatrix)):
writing = ""
for j in range(len(CalcMatrix)):
if str(CalcMatrix[i][j]) == "0.0":
writing = writing + " " + "?"
else:
writing = writing + " " + str(CalcMatrix[i][j])
### Add break line
writing = writing + " " + "\n"
outfile.write(writing)
outfile.close()
for i in range(len(BackboneDisp)):
line="%3s %4s %3s %3s"%(BackboneDisp[i][0],round(BackboneDisp[i][1],1),BackboneDisp[i][2],BackboneDisp[i][3])
backboneoutfile.write(line+'\n')
backboneoutfile.close()
###Make gnuplot plot file
gnuoutfile.write("reset" + "\n")
gnuoutfile.write("cd " + "'" + os.getcwd() + "'" + "\n")
gnuoutfile.write("\n")
gnuoutfile.write("#Title hacks \\n is newline, and 0,1 is x,y offset adjustment" + "\n")
gnuoutfile.write('set title "Protein ' + str(atom) + ' Displacement matrix map \\n ResRes min. ' + str(mindist) + ' Ang, ' + 'Delta min. ' + str(mindelta) + ' Ang" 0,1' + "\n")
gnuoutfile.write("# x is column" + "\n")
gnuoutfile.write("set xlabel 'Res nr. for " + str(molecule2) +"'"+ "\n")
gnuoutfile.write("# y is row" + "\n")
gnuoutfile.write("set ylabel 'Res nr. for " + str(molecule1) +"'"+ "\n")
gnuoutfile.write("\n")
gnuoutfile.write("#set xrange [300:550]; set yrange [0:400]" + "\n")
gnuoutfile.write("#set xtics 50" + "\n")
gnuoutfile.write("#set ytics 50" + "\n")
gnuoutfile.write("#set mxtics 5" + "\n")
gnuoutfile.write("#set mytics 5" + "\n")
gnuoutfile.write("set size ratio 1" + "\n")
gnuoutfile.write("unset key" + "\n")
gnuoutfile.write("\n")
gnuoutfile.write("set cbrange [-30:30]" + "\n")
gnuoutfile.write("set palette defined (-30 'blue', 0 'white', 30 'red')" + "\n")
gnuoutfile.write("set pm3d map" + "\n")
gnuoutfile.write("\n")
gnuoutfile.write("#set term postscript eps enhanced color" + "\n")
gnuoutfile.write('#set output "'+ filename + '.eps"' + "\n")
gnuoutfile.write("set term png" + "\n")
gnuoutfile.write('set output "'+ filename + '.png"' + "\n")
gnuoutfile.write("splot '" + str(filename+".txt") + "' matrix" + "\n")
gnuoutfile.write("\n")
gnuoutfile.write("#For the backbone displacement"+"\n")
gnuoutfile.write("\n")
gnuoutfile.write('set title "Protein ' + str(atom) + ' Backbone displacement" 0,1' + "\n")
gnuoutfile.write("set xlabel 'Residue number'" + "\n")
gnuoutfile.write("set ylabel '" +str(atom) + " displacement (Ang)'" + "\n")
gnuoutfile.write("\n")
gnuoutfile.write("#set xrange [0:550]; set yrange [0:40]" + "\n")
gnuoutfile.write("#set xtics 50" + "\n")
gnuoutfile.write("#set ytics 10" + "\n")
gnuoutfile.write("#set mxtics 5" + "\n")
gnuoutfile.write("#set mytics 5" + "\n")
gnuoutfile.write("set size ratio 0.75" + "\n")
gnuoutfile.write("unset key" + "\n")
gnuoutfile.write("\n")
gnuoutfile.write("#set term postscript eps enhanced color" + "\n")
gnuoutfile.write('#set output "'+ filename + '-backbone.eps"' + "\n")
gnuoutfile.write("set term png" + "\n")
gnuoutfile.write('set output "'+ filename + '-backbone.png"' + "\n")
gnuoutfile.write("plot '" + str(filename+"-backbone.txt") + "' using 1:2 title 'Backbone displacement' with lines" + "\n")
gnuoutfile.close()
###Create stick residue objects
for i in range(len(MaxNegDist)):
name = str(i) + "_" + str(round(MaxNegDist[i][0],1))+"_"+shortaa(str(MaxNegDist[i][5]))+str(MaxNegDist[i][1])+shortaa(str(MaxNegDist[i][6]))+str(MaxNegDist[i][2])
selection = str(molecule1)+" and resi "+str(MaxNegDist[i][1]) + "+"+str(MaxNegDist[i][2])+" or "+str(molecule2)+" and resi "+str(MaxNegDist[i][2])
#print selection
cmd.create(name, selection)
if showsticks=='yes' or showsticks=='y':
cmd.show("sticks", name)
for i in range(len(MaxPosDist)):
name = str(i) + "_" + str(round(MaxPosDist[i][0],1))+"_"+shortaa(str(MaxPosDist[i][5]))+str(MaxPosDist[i][1])+shortaa(str(MaxPosDist[i][6]))+str(MaxPosDist[i][2])
selection = str(molecule1)+" and resi " + str(MaxPosDist[i][1])+"+" + str(MaxPosDist[i][2])+" or " + str(molecule2)+" and resi "+str(MaxPosDist[i][2])
#print selection
cmd.create(name, selection)
if showsticks=='yes' or showsticks=='y':
cmd.show("sticks", name)
cmd.extend("dispmap",dispmap)
def create_nXn_matrix(n):
return [[0.0 for x in range(n)] for x in range(n)]
def distance(array1, array2, i, j):
i = int(i); j = int(j)
dist = sqrt((array1[i][0] - array2[j][0])**2 + (array1[i][1] - array2[j][1])**2 + (array1[i][2] - array2[j][2])**2)
return dist
def Coord(Input):
print cmd.get_atom_coords(Input)
cmd.extend("Coord",Coord)
def replace_words(text, word_dic):
rc = re.compile('|'.join(map(re.escape, word_dic)))
def translate(match):
return word_dic[match.group(0)]
return rc.sub(translate, text)
def shortaa(longaa):
aa_dic = {'ARG':'R','HIS':'H','LYS':'K',
'ASP':'D','GLU':'E',
'SER':'S','THR':'T','ASN':'N','GLN':'Q',
'CYS':'C','SEC':'U','GLY':'G','PRO':'P',
'ALA':'A','ILE':'I','LEU':'L','MET':'M','PHE':'F','TRP':'W','TYR':'Y','VAL':'V'}
return(replace_words(longaa, aa_dic))
def cysb62(aa):
#BLOSUM62 cys mutation
# C S T P A G N D E Q H R K M I L V F Y W
#C9 -1 -1 -3 0 -3 -3 -3 -4 -3 -3 -3 -3 -1 -1 -1 -1 -2 -2 -2
b62_dic = {'R':'-3','H':'-3','K':'-1',
'D':'-3','E':'-4',
'S':'-1','T':'-1','N':'-3','Q':'-3',
'C':'9','U':'9','G':'-3','P':'-3',
'A':'0','I':'-1','L':'-1','M':'-1','F':'-2','W':'-2','Y':'-2','V':'-1'}
return(replace_words(aa, b62_dic))
def pam250(aa):
# A R N D C Q E G H I L K M F P S T W Y V
#C 2 1 1 1 52 1 1 2 2 2 1 1 1 1 2 3 2 1 4 2
# Mutation probability matrix for the evolutionary distance of 250 PAMs.
# To simplify the appearance, the elements are shown multiplied by 100.
# In comparing two sequences of average amino acid frequency at this evolutionary distance,
# there is a 13% probability that a position containing Ala in the first sequence will contain Ala in the second.
# There is a 3% chance that it will contain Arg, and so forth.
# (Adapted from Figure 83. Atlas of Protein Sequence and Structure, Suppl 3, 1978, M.O. Dayhoff, ed. National Biomedical Research Foundation, 1979.)
pam250_dic = {'R':'1','H':'2','K':'1',
'D':'1','E':'1',
'S':'3','T':'2','N':'1','Q':'1',
'C':'52','U':'52','G':'2','P':'2',
'A':'2','I':'2','L':'1','M':'1','F':'1','W':'1','Y':'4','V':'2'}
return(replace_words(aa, pam250_dic))
References
For EPR considerations
Conformation of T4 Lysozyme in Solution. Hinge-Bending Motion and the Substrate-Induced Conformational Transition Studied by Site-Directed Spin Labeling
Hassane S. Mchaourab, Kyoung Joon Oh, Celia J. Fang, and Wayne L. Hubbell
Biochemistry 1997, 36, 307-316
Probing Single-Molecule T4 Lysozyme Conformational Dynamics by Intramolecular Fluorescence Energy Transfer
Yu Chen, Dehong Hu, Erich R. Vorpagel, and H. Peter Lu
J. Phys. Chem. B 2003, 107, 7947-7956
For FRET pair selection and considerations
Fluorescent probes and bioconjugation chemistries for single-molecule fluorescence analysis of biomolecules
Achillefs N. Kapanidisa and Shimon Weiss
Journal of chemical physics VOLUME 117, Number 24 22 December 2002
For inspiration to DisplacementMap. Fig: 6, Difference-distance matrix for the difference in CA-CA distances.
Structure of a Hinge-bending Bacteriophage T4 Lysozyme Mutant, Ile3 -> Pro
M. M. Dixon, H. Nicholsont, L. Shewchuk W. A. Baase and B. W. Matthews1
J. Mol. Biol. (1992) 227. 917-933