Difference between revisions of "Dehydron"
(→Usage) |
|||
| Line 44: | Line 44: | ||
Another two control the dehydron detection. | Another two control the dehydron detection. | ||
| − | * Desolvatation sphere radius: this parameter controls the radius of the two spheres | + | * Desolvatation sphere radius: this parameter controls the radius of the two spheres centred at the Cα carbon of the donor and acceptor residues. A dehydron is defined by the number of "wrappers" inside this two spheres.<br> |
| − | * Min wrappers: a hydrogen bond surrounded with less "wrappers" than "min_wrappers" is a dehydron. Setting this parameter to a "high" value, something like 100, will return all main chain hydrogen bonds (according to the angle range and max distance parameters). | + | * Min wrappers: a hydrogen bond surrounded with less "wrappers" than "min_wrappers" is a dehydron. Setting this parameter to a "high" value, something like 100, will return all main chain hydrogen bonds (according to the angle range and max distance parameters). The default value (19) was taken from a statistical analysis of ~7400 high-quality proteins form the Protein Data Bank. The frequency of wrappers in this set of proteins closely follow a Gaussian distribution with a mean of 27 and a standard deviation of 8 (27-8 = 19). |
| − | A wrapper is defined as a carbon atom not bonded directly to an oxygen or nitrogen atom, i.e. a non-polar carbon atom. The | + | A wrapper is defined as a carbon atom not bonded directly to an oxygen or nitrogen atom, i.e. a non-polar carbon atom. The plug-in count as wrappers any non-polar carbon from any protein chain, organic ligand or other type of molecule, if the atoms belong "selection" (see below). This means that if you have, for example, a dimeric protein you will probably get different results for the dimer and for the isolated monomers. Instead, if you upload two (or more) different files the results will be independent because the plug-in does not count atoms from other objects |
| − | * Selection: This parameter allows the user to select which part of system is used to calculate dehydrons. This parameter is useful, for example, to calculate dehydronds for different objects independently or to easily calculate dehydrons with and without an organic ligand. The default selection is "all". Compute dehydrons for an | + | * Selection: This parameter allows the user to select which part of system is used to calculate dehydrons. This parameter is useful, for example, to calculate dehydronds for different objects independently or to easily calculate dehydrons with and without an organic ligand. The default selection is "all". Compute dehydrons for an specific selection is equivalent to delete all but the selected atoms and then compute dehydrons. |
== Acknowledgement == | == Acknowledgement == | ||
The H-bond detection code is based on the list_mc_hbonds.py script from Robert L. Campbell http://pldserver1.biochem.queensu.ca/~rlc/work/pymol/ | The H-bond detection code is based on the list_mc_hbonds.py script from Robert L. Campbell http://pldserver1.biochem.queensu.ca/~rlc/work/pymol/ | ||
| − | == | + | == Change log == |
* 2012-01-14 (Version 1.0) | * 2012-01-14 (Version 1.0) | ||
| Line 75: | Line 75: | ||
*# Wrappers were not correctly counted for structures with hydrogen atoms. | *# Wrappers were not correctly counted for structures with hydrogen atoms. | ||
Thanks Shafqat Rasool for reporting the bug. | Thanks Shafqat Rasool for reporting the bug. | ||
| + | |||
| + | *2013-07-19 (Version 2.0) | ||
| + | *# The plug-in was renamed to wrappy. | ||
| + | *# Wrappy reports, now, the hydrogen bonds per residue and the wrappers per residue as z-scores. The values of the mean and standard deviation, necessary to compute zscores, where taken from the analysis of ~7400 high quality X-ray proteins from the PDB. | ||
== References == | == References == | ||
Revision as of 16:15, 19 July 2013
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/dehydron.py |
| Author(s) | Osvaldo Martin |
| License | GPL |
| This code has been put under version control in the project Pymol-script-repo | |
Introduction
A dehydron calculator plugin for PyMOL. This plugin calculates dehydrons and display them onto the protein structure.
A dehydron is a protein main chain hydrogen bond incompletely shielded from water attack. Dehydrons are sticky, since they promote the removal of surrounding water through protein associations or ligand binding. Dehydrons are less conserved than other structural motifs, hence identification of dehydrons could help to increase specificity during the rational drug design process.
For a brief introduction to the dehydron concept, please read wikipedia.
Installation
Linux
This plugin is ready "out-of-box" for Linux users through the project Pymol-script-repo
Windows
This plugin is ready "out-of-box" for Windows users through the project Pymol-script-repo
Mac OsX
This plugin have not been tested on a Mac OsX machine, but it should work.
Usage
The plugin can be accessed using the following command:
dehydron [ selection [, angle_range [, max_distance [, desolv [, min_wrappers ]]]]]
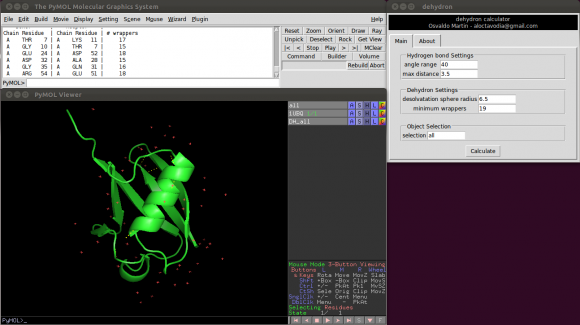
Or using a graphical environment (see accompanying figure)
There are five parameters the user can change:
Two of them control the hydrogen bonds detection.
- Angle range: deviation in degrees from the optimal hydrogen bond angle (C=0 N-H).
- Max distance: maximum donor-acceptor distance.
Another two control the dehydron detection.
- Desolvatation sphere radius: this parameter controls the radius of the two spheres centred at the Cα carbon of the donor and acceptor residues. A dehydron is defined by the number of "wrappers" inside this two spheres.
- Min wrappers: a hydrogen bond surrounded with less "wrappers" than "min_wrappers" is a dehydron. Setting this parameter to a "high" value, something like 100, will return all main chain hydrogen bonds (according to the angle range and max distance parameters). The default value (19) was taken from a statistical analysis of ~7400 high-quality proteins form the Protein Data Bank. The frequency of wrappers in this set of proteins closely follow a Gaussian distribution with a mean of 27 and a standard deviation of 8 (27-8 = 19).
A wrapper is defined as a carbon atom not bonded directly to an oxygen or nitrogen atom, i.e. a non-polar carbon atom. The plug-in count as wrappers any non-polar carbon from any protein chain, organic ligand or other type of molecule, if the atoms belong "selection" (see below). This means that if you have, for example, a dimeric protein you will probably get different results for the dimer and for the isolated monomers. Instead, if you upload two (or more) different files the results will be independent because the plug-in does not count atoms from other objects
- Selection: This parameter allows the user to select which part of system is used to calculate dehydrons. This parameter is useful, for example, to calculate dehydronds for different objects independently or to easily calculate dehydrons with and without an organic ligand. The default selection is "all". Compute dehydrons for an specific selection is equivalent to delete all but the selected atoms and then compute dehydrons.
Acknowledgement
The H-bond detection code is based on the list_mc_hbonds.py script from Robert L. Campbell http://pldserver1.biochem.queensu.ca/~rlc/work/pymol/
Change log
- 2012-01-14 (Version 1.0)
- First public version was released and put under version control. In the project, Pymol-script-repo.
- 2012-01-28 (Version 1.1)
- Minor changes in the code most of them not visible for the end-user.
- 2012-02-28 (Version 1.5)
- The code was cleaned (e.g. remove global variables and other ugly stuff)
- The code was made available as a PyMOL command
- Better support for multiple objects
All features in this version and most of the code was provided by Thomas Holder, thanks Thomas! :-)
- 2012-03-14 (Version 1.6)
- Representation is not changed to "cartoon" after each calculation
- Total control over the selection from which dehydrons are calculated
- 2013-03-26 (Version 1.7)
- Wrappers were not correctly counted for structures with hydrogen atoms.
Thanks Shafqat Rasool for reporting the bug.
- 2013-07-19 (Version 2.0)
- The plug-in was renamed to wrappy.
- Wrappy reports, now, the hydrogen bonds per residue and the wrappers per residue as z-scores. The values of the mean and standard deviation, necessary to compute zscores, where taken from the analysis of ~7400 high quality X-ray proteins from the PDB.
References
Citation for Dehydrons:
Fernández A. and Scott R.; "Adherence of packing defects in soluble proteins", Phys. Rev. Lett. 91, 018102 (2003).
Fernández A., Rogale K., Scott R. and Scheraga H.A.; "Inhibitor design by wrapping packing defects in HIV-1 proteins", PNAS, 101, 11640-45 (2004).
Fernández A. "Transformative Concepts for Drug Design: Target Wrapping" (ISBN 978-3642117916), Springer-Verlag, Berlin, Heidelberg (2010).