Difference between revisions of "Colorbydisplacement"
| Line 47: | Line 47: | ||
== Example Pymol Script == | == Example Pymol Script == | ||
<syntaxhighlight lang="python"> | <syntaxhighlight lang="python"> | ||
| − | + | import colorbydisplacement | |
| − | |||
| − | |||
| − | |||
| − | |||
| + | fetch 1HP1, async=0 | ||
| + | fetch 1HPU, async=0 | ||
| + | |||
hide everything | hide everything | ||
### Select asymmetric units from pdb file | ### Select asymmetric units from pdb file | ||
| − | create O5NT, / | + | create O5NT, /1HP1//A |
| − | create C5NT, / | + | create C5NT, /1HPU//C |
| − | delete | + | delete 1HP1 |
| − | delete | + | delete 1HPU |
| − | + | ||
| − | |||
show cartoon, O5NT | show cartoon, O5NT | ||
show cartoon, C5NT | show cartoon, C5NT | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| + | ColorByDisplacementAll O5NT, C5NT, super1=resi 26-355, super2=resi 26-355, doColor=t, doAlign=t | ||
| + | |||
set_view (\ | set_view (\ | ||
0.094686687, -0.390707940, 0.915631354,\ | 0.094686687, -0.390707940, 0.915631354,\ | ||
| Line 84: | Line 72: | ||
231.830673218, 330.050415039, -20.000000000 ) | 231.830673218, 330.050415039, -20.000000000 ) | ||
</syntaxhighlight> | </syntaxhighlight> | ||
| − | |||
== Python Code == | == Python Code == | ||
Revision as of 11:14, 1 December 2011
Acknowledgement
This pymol script is made by Troels Emtekær Linnet.
This script is based on the scaffold from ColorByRMSD. Peace love and harmony goes to Shivender Shandilya and Jason Vertrees.
Introduction
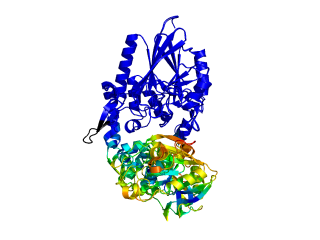
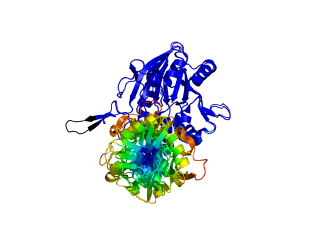
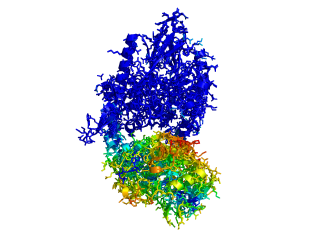
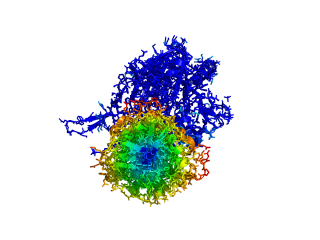
This script allows you to color two structures by distance displacement between an Open and Closed form of a protein, as calculated by PyMol's internal distance command. The pairwise distance is calculated between, C-alpha or all-atoms. The distance displacement values are stored as B-factors of these residues, which are colored by a rainbow color spectrum, with blue specifying minimum and red indicating maximum.
Code
Do keep in mind, all original B-factors values are overwritten!
There exist two versions.
ColorByDisplacementCA is quick and is between CA atoms. Ideal for helices representation.
ColorByDisplacementAll is between All atoms in residues and is quite slow => 3-5 mins for a run. Ideal for sticks representation.
You have to specify which residues should be used in the alignment procedure, or it will take all residues as standard
V.2 is implemented the 2011.01.06 - Due to a bug in coloring.
Bug in code
A bug in the boolean operator of the spectrum command has been found. This versions work for version 1.3 Educational product.
For other versions of pymol, try to change (comment/uncomment) the cmd.spectrum line.
The other spectrum line works for Open-Source PyMOL 1.2r3pre, Incentive product
Examples
ColorByDisplacementCA O5NT, C5NT, super1=resi 26-355, super2=resi 26-355, doColor=t, doAlign=t
ColorByDisplacementAll O5NT, C5NT, super1=resi 26-355, super2=resi 26-355, doColor=t, doAlign=t
ColorByDisplacementCA O5NT, C5NT, super1=resi 26-355, super2=resi 26-355, doColor=t, doAlign=t, AlignedWhite='no'
ColorByDisplacementAll O5NT, C5NT, super1=resi 26-355, super2=resi 26-355, doColor=t, doAlign=t, AlignedWhite='no'
ColorByDisplacementCA O5NT, C5NT, resi 26-355, resi 26-355
ColorByDisplacementAll O5NT, C5NT, resi 26-355, resi 26-355
Dark blue is low displacement, higher displacements are in orange/yellow/red.
Residues used for alignment is colored white. Can be turned off in top of algorithm.
Residues not in both pdb files is colored black
Example Pymol Script
import colorbydisplacement
fetch 1HP1, async=0
fetch 1HPU, async=0
hide everything
### Select asymmetric units from pdb file
create O5NT, /1HP1//A
create C5NT, /1HPU//C
delete 1HP1
delete 1HPU
show cartoon, O5NT
show cartoon, C5NT
ColorByDisplacementAll O5NT, C5NT, super1=resi 26-355, super2=resi 26-355, doColor=t, doAlign=t
set_view (\
0.094686687, -0.390707940, 0.915631354,\
0.809000611, -0.505792081, -0.299485058,\
0.580131471, 0.769104064, 0.268191338,\
0.000000000, 0.000000000, -280.940521240,\
26.240486145, 46.146961212, 21.702068329,\
231.830673218, 330.050415039, -20.000000000 )
Python Code
This code has been put under version control. In the project, Pymol-script-repo.
For a color coded view:
https://github.com/Pymol-Scripts/Pymol-script-repo/blob/master/colorbydisplacement.py
See the raw code or download manually, by right clicking the following link here -> Save as: colorbydisplacement.py
https://raw.github.com/Pymol-Scripts/Pymol-script-repo/master/colorbydisplacement.py