Difference between revisions of "Cheshift"
m |
|||
| Line 40: | Line 40: | ||
== Using the Plug-in == | == Using the Plug-in == | ||
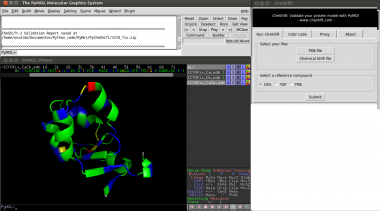
| − | 1) Launch PyMOL and select " | + | 1) Launch PyMOL and select "Cheshift" from the Plug-ins menu. <br> |
| − | 2) Select a PDB file of your protein model (using the CheShift plug-in). | + | 2) Select a PDB file of your protein model (using the CheShift plug-in menu).<br> |
| − | 3) Select a file with the experimental | + | 3) Select a file with the experimental chemical shift values.<br> |
| − | 4) Click the "Submit" button. | + | 4) Click the "Submit" button.<br> |
5) Wait until results are displayed (this could take a few minutes, depending | 5) Wait until results are displayed (this could take a few minutes, depending | ||
on all existing request on the server. Analyzing 20 conformations of a 100 | on all existing request on the server. Analyzing 20 conformations of a 100 | ||
| − | residues protein takes ~4 minutes). | + | residues protein takes ~4 minutes).<br> |
| − | Note: | + | <b>Note:</b> |
| − | * If more than 20 conformers are uploaded only the 20 first will be analyzed | + | * If more than 20 conformers are uploaded only the 20 first will be analyzed. |
| − | * If the PDB file has more than one chain only the first one will be analyzed | + | * If the PDB file has more than one chain only the first one will be analyzed. |
| − | * The PDB file must not have missing residues | + | * The PDB file must not have missing residues. |
| − | * Missing observed <sup>13</sup>C<sup>α</sup> chemical shifts are tolerated | + | * Missing observed <sup>13</sup>C<sup>α</sup> chemical shifts are tolerated. |
* The observed <sup>13</sup>C<sup>α</sup> chemical shifts values should be in BMRB-Star format (only | * The observed <sup>13</sup>C<sup>α</sup> chemical shifts values should be in BMRB-Star format (only | ||
| − | the <sup>13</sup>C<sup>α</sup> are needed, the others nuclei could be present but are ignored) | + | the <sup>13</sup>C<sup>α</sup> are needed, the others nuclei could be present but are ignored). |
you can use a complete BMRB file or just a file with only the experimental chemical shifts. In the latter case you can use this format: | you can use a complete BMRB file or just a file with only the experimental chemical shifts. In the latter case you can use this format: | ||
| Line 97: | Line 97: | ||
CheShift plug-in is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License. A complete copy of the GNU General Public License can be accessed here http://www.gnu.org/licenses/. | CheShift plug-in is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License. A complete copy of the GNU General Public License can be accessed here http://www.gnu.org/licenses/. | ||
| − | CheShift Server (www.cheshift.com) is available free of charge ONLY for academic use. | + | CheShift Server ([http://www.cheshift.com www.cheshift.com]) is available free of charge ONLY for academic use. |
Revision as of 03:30, 14 February 2012
| Type | PyMOL Plugin |
|---|---|
| Download | plugins/cheshift.py |
| Author(s) | Osvaldo Martin |
| License | GPL |
| This code has been put under version control in the project Pymol-script-repo | |
Description
The differences between observed and predicted 13Cα chemical shifts can be used as a sensitive probe with which to detect possible local flaws in protein structures. For this reason exist CheShift, a Web server for protein structure validation.
This plug-in provides a way to use PyMOL to validate a protein model using observed chemical shifts. All computations run on CheShift server, and the results are retrieved to PyMOL, hence an Internet connection is needed.
Version
The current version of this plug-in is 1.0
Installation
Linux
1) The plug-in can be downloaded from here [Pymol-script-repo]
2) You should install mechanize, a python module. This module is available from the repositories of the main Linux distributions. Just use your default package manager (or command line) to install it. In Ubuntu/Debian the package name is "python-mechanize".
Windows
This plug-in have not been extensively tested on Windows machines, but it passed all the test I have done...
1) The plug-in can be downloaded from here [Pymol-script-repo]
2) You should install mechanize, a python module. In order to do that, download this unzip and copy the mechanize folder were you have installed PyMOL, usually is C:\Program Files\DeLano Scientific\PyMOL\modules\pmg_tk\startup
Mac OsX
This plug-in have not been tested on a Mac OsX machine, but it should work...
1) The plug-in can be downloaded from here [Pymol-script-repo]
2) You should install mechanize, a python module. In order to do that download this unzip and copy the mechanize folder were you have installed PyMOL. For the X11/Hybrid version, the location is probably: PyMOLX11Hybrid.app/pymol/modules/pmg_tk/startup
Using the Plug-in
1) Launch PyMOL and select "Cheshift" from the Plug-ins menu.
2) Select a PDB file of your protein model (using the CheShift plug-in menu).
3) Select a file with the experimental chemical shift values.
4) Click the "Submit" button.
5) Wait until results are displayed (this could take a few minutes, depending
on all existing request on the server. Analyzing 20 conformations of a 100
residues protein takes ~4 minutes).
Note:
- If more than 20 conformers are uploaded only the 20 first will be analyzed.
- If the PDB file has more than one chain only the first one will be analyzed.
- The PDB file must not have missing residues.
- Missing observed 13Cα chemical shifts are tolerated.
- The observed 13Cα chemical shifts values should be in BMRB-Star format (only
the 13Cα are needed, the others nuclei could be present but are ignored).
you can use a complete BMRB file or just a file with only the experimental chemical shifts. In the latter case you can use this format:
1 21 MET N N 121.0 0.25 1
2 21 MET H H 8.36 0.01 1
3 21 MET CA C 55.9 0.20 1
4 21 MET HA H 4.39 0.01 1
5 21 MET CB C 31.8 0.20 1
6 21 MET HB2 H 2.15 0.01 2
7 21 MET HG2 H 2.54 0.01 2
8 21 MET HE H 2.09 0.01 1
9 21 MET C C 176.6 0.10 1
10 22 GLY N N 111.4 0.25 1
11 22 GLY H H 8.55 0.01 1
12 22 GLY CA C 44.9 0.20 1
13 22 GLY HA2 H 4.15 0.01 2
14 22 GLY HA3 H 3.91 0.01 2
15 22 GLY C C 173.2 0.10 1
or this format:
1 1 1 SER HA H 4.44 0.005 1
2 1 1 SER HB2 H 3.85 0.005 2
3 1 1 SER HB3 H 3.88 0.005 2
4 1 1 SER C C 168.6 0.2 1
5 1 1 SER CA C 58.4 0.2 1
6 1 1 SER CB C 63.8 0.2 1
7 2 2 ALA H H 8.23 0.005 1
8 2 2 ALA HA H 4.87 0.005 1
9 2 2 ALA HB H 1.37 0.005 1
10 2 2 ALA C C 174.5 0.2 1
11 2 2 ALA CA C 51.7 0.2 1
12 2 2 ALA CB C 23.3 0.2 1
13 2 2 ALA N N 120.2 0.2 1
Proxy Configuration
If you are behind a proxy the CheShift plug-in will try to correctly guess your proxy configuration, but this is a tricky business and many things could fail. In that scenario you will be prompted to manually set your proxy settings. In case you manually set the proxy configuration the plug-in will save your proxy settings and next time it will attempt to use those saved settings to connect to the Internet.
License
CheShift plug-in is free software: you can redistribute it and/or modify it under the terms of the GNU General Public License. A complete copy of the GNU General Public License can be accessed here http://www.gnu.org/licenses/.
CheShift Server (www.cheshift.com) is available free of charge ONLY for academic use.
Citation
Martin O.A. Vila J.A. and Scheraga H.A. CheShift-2: Graphic validation of protein structures. Bioinformatics 2012 (submitted)
References
Martin O.A. Vila J.A. and Scheraga H.A. (2012). CheShift-2: Graphic validation of protein structures. Bioinformatics 2012 (submitted).
Vila J.A. Arnautova Y.A. Martin O.A. and Scheraga, H.A. (2009). Quantum-mechanics-derived 13C chemical shift server (CheShift) for protein structure validation. PNAS, 106(40), 16972-16977.
Vila, J.A. and Scheraga H.A. (2009). Assessing the accuracy of protein structures by quantum mechanical computations of 13Cα chemical shifts. Accounts of chemical research, 42(10), 1545-53.