Difference between revisions of "APBS"
| Line 3: | Line 3: | ||
[http://apbs.sourceforge.net APBS], the Adaptive Poisson-Boltzmann Solver, is a [http://www.oreilly.com/openbook/freedom/ freely] available macromolecular electrostatics calculation program released under the [http://www.gnu.org/copyleft/gpl.html GPL]. It is a cost-effective but uncompromised alternative to [http://trantor.bioc.columbia.edu/grasp/ GRASP], and it can be used within pymol. Pymol can display the results of the calculations as an electrostatic potential molecular surface. | [http://apbs.sourceforge.net APBS], the Adaptive Poisson-Boltzmann Solver, is a [http://www.oreilly.com/openbook/freedom/ freely] available macromolecular electrostatics calculation program released under the [http://www.gnu.org/copyleft/gpl.html GPL]. It is a cost-effective but uncompromised alternative to [http://trantor.bioc.columbia.edu/grasp/ GRASP], and it can be used within pymol. Pymol can display the results of the calculations as an electrostatic potential molecular surface. | ||
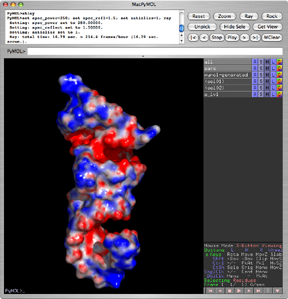
| − | PyMol currently supports the '''APBS plugin''' written by Michael Lerner. This plugin makes it possible to run APBS from within PyMOL, and then display the results as a color-coded electrostatic surface (units | + | PyMol currently supports the '''APBS plugin''' written by Michael Lerner. This plugin makes it possible to run APBS from within PyMOL, and then display the results as a color-coded electrostatic surface (units <math>K_bT/e_c</math>) in the molecular display window (as with the image to the right). See [http://apbs.wustl.edu/MediaWiki/index.php/APBS_electrostatics_in_PyMOL the APBS wiki] for more details, including instructions on how to download, install and use the plugin. |
'''Nucleic acids may prove problematic for the apbs plugin.''' If so, use the [http://pdb2pqr.sourceforge.net/ pdb2pqr] command-line tool to create a pqr file manually, instead of using the plugin to generate it. Then direct the APBS GUI on the [http://www-personal.umich.edu/~mlerner/PyMOL/images/main.png main menu] to read the pqr file you '''externally generated.''' | '''Nucleic acids may prove problematic for the apbs plugin.''' If so, use the [http://pdb2pqr.sourceforge.net/ pdb2pqr] command-line tool to create a pqr file manually, instead of using the plugin to generate it. Then direct the APBS GUI on the [http://www-personal.umich.edu/~mlerner/PyMOL/images/main.png main menu] to read the pqr file you '''externally generated.''' | ||
Revision as of 17:42, 1 April 2009
Introduction
APBS, the Adaptive Poisson-Boltzmann Solver, is a freely available macromolecular electrostatics calculation program released under the GPL. It is a cost-effective but uncompromised alternative to GRASP, and it can be used within pymol. Pymol can display the results of the calculations as an electrostatic potential molecular surface.
PyMol currently supports the APBS plugin written by Michael Lerner. This plugin makes it possible to run APBS from within PyMOL, and then display the results as a color-coded electrostatic surface (units ) in the molecular display window (as with the image to the right). See the APBS wiki for more details, including instructions on how to download, install and use the plugin.
Nucleic acids may prove problematic for the apbs plugin. If so, use the pdb2pqr command-line tool to create a pqr file manually, instead of using the plugin to generate it. Then direct the APBS GUI on the main menu to read the pqr file you externally generated.
Required Dependencies
APBS and its dependencies like pdb2pqr and maloc are freely available under the GPL. The author of the software however asks that users register with him to aid him in obtaining grant funding.
Installing the Dependencies on OS X
- First, register your use of the software. This will keep everyone happy.
- Second, if you don't already have the fink package management system, now is a good time to get it. Here is a quick-start set of instructions for getting X-windows, compilers, and fink all installed.
- Once you are up and going, activate the unstable branch in fink, and then issue the commands
fink self-update
fink install apbs
or if you want to use the multi-processor version, issue
fink self-update
fink install apbs
Then install the X-windows based version of pymol using the command
fink install pymol-py25
Note that the fink version of pymol already has the latest version of the APBS plugin. You are set to go!
Further details, as well as screen shots, are given elsewhere in this wiki.
Installing the Dependencies on Linux
From Scratch
Note that this tutorial assumes you're using the bash shell and have root privileges
-
Obtain APBS and MALOC from...
APBS = http://apbs.sourceforge.net (currently 0.4)
MALOC = http://www.fetk.org/codes/maloc/index.html#download (currently 0.1-2)
- Set up some environment variables & directories (temporary for building)
$ export FETK_SRC=/<building directory>/temp_apbs $ export FETK_PREFIX=/usr/local/apbs-0.4.0 (or wherever you want it to live) $ export FETK_INCLUDE=${FETK_PREFIX}/include $ export FETK_LIBRARY=${FETK_PREFIX}/lib $ mkdir -p ${FETK_SRC} ${FETK_INCLUDE} ${FETK_LIBRARY}
- Unpack the source packages
$ cd ${FETK_SRC} $ gzip -dc maloc-0.1-2.tar.gz | tar xvf - $ gzip -dc apbs-0.4.0.tar.gz | tar xvf -
- Compile MALOC
$ cd ${FETK_SRC}/maloc $ ./configure --prefix=${FETK_PREFIX}
If everything went well, then
$ make; make install - Go get a coffee. Compilation/installation takes about 15 minutes on a 3GHz computer with 1GB of RAM.
- Now on to compiling APBS itself
$ cd ${FETK_SRC}/apbs-0.4.0 $ ./configure --prefix=${FETK_PREFIX}
If all goes well:
$ make all; make install - No time for coffee. Takes about 5 minutes on that fast computer.
- There will now be an APBS binary at
/usr/local/apbs-0.4.0/bin/i686-intel-linux/apbs
- Make appropriate links
$ ln -s /usr/local/apbs-0.4.0/bin/i686-intel-linux/apbs /usr/local/bin/apbs
- Get rid of /<building directory dir>/temp_apbs
- Open PyMOL and make sure that the APBS plugin points to /usr/local/bin/apbs
- Rock and or Roll.
Pre-Packaged
RPMs
A variety of RPMs are available from the APBS downloads website. Again, please register your use of the software if you have not yet done so.
Debian packages
For ubuntu and other debian linux distributions, probably the simplest thing is to download a promising looking rpm, convert it with the program alien, and then install the newly generated debian package with the command
sudo dpkg -i apbs*.deb
Gentoo
You have to install apbs and pdb2pqr. Both are masked via keywords atm. Type as root:
echo sci-chemistry/pdb2pqr >> /etc/portage/package.keywords
echo sci-chemistry/apbs >> /etc/portage/package.keywords
emerge -av sci-chemistry/apbs sci-chemistry/pdb2pqr
Troubleshooting
- If the B-factor is then APBS doesn't properly read in the PDB file and thus outputs garbage (or dies). To fix this, set all b factors to be less than 100. The problem stems from how to parse a PDB file. The PDB file originally was written when most people used FORTRAN programs, and so the file format was specified by columns, not by the more modern comma separated value format we tend to prefer today. For the latest on the PDB format see the new PDB format docs.
alter all, b=min(b,99.9)
- APBS has problems, sometimes, in reading atoms with alternate conformations. You can remove the alternate locations with a simple script removeAlt.
- ObjectMapLoadDXFile-Error: as of this writing (9-23-2008) a known problem exists, and the Baker lab is working on it.