Difference between revisions of "Visualizing a computed structure - a commented example"
Qqqqqqqqq9 (talk | contribs) |
Qqqqqqqqq9 (talk | contribs) |
||
| Line 20: | Line 20: | ||
6. Use the script to modify your own pdb-file. | 6. Use the script to modify your own pdb-file. | ||
| + | |||
| + | [[Category:Tutorials|Visualizing a computed structure - a commented example]] | ||
Revision as of 07:35, 12 September 2008
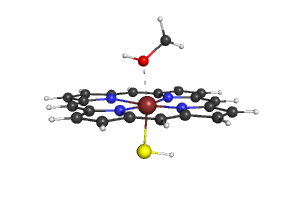
1. Obtain the data, the following structure was published in http://pubs.acs.org/cgi-bin/abstract.cgi/jacsat/2000/122/i37/abs/ja991878x.html in the Supporting Information
2. Save File.xyz.tar and untar it.
3. a) convert it to pdb with Openbabel (http://openbabel.org)
babel file.xyz file.pdb
or 3. b)
Open it with avogadro (http://avogadro.openmolecules.net) and save it as pdb. Avogadro allows you to adjust the coordinate sytem, which saves some work if you want to visualize several similar structures.
4. Save the following pymolscript Script.pml.tar and untar it to script.pml. Open it with an editor and adjust the Path_To_The_PDB-File. Open pymol and run the script with "@PATH_Of_The_Script/script.pml.
5. You will get the following image, you can save it with "png filename.png"
6. Use the script to modify your own pdb-file.