Radius of gyration
Jump to navigation
Jump to search
The printable version is no longer supported and may have rendering errors. Please update your browser bookmarks and please use the default browser print function instead.
|
Included in psico | |
| Module | psico.querying#gyradius |
|---|---|
This script does calcualate the radius of gyration of a molecule.
Thanks to Tsjerk Wassenaar for posting this script on the pymol-users mailing list!
The function was added to psico as "gyradius".
from pymol import cmd
import math
def rgyrate(selection='(all)', quiet=1):
'''
DESCRIPTION
Radius of gyration
USAGE
rgyrate [ selection ]
'''
try:
from itertools import izip
except ImportError:
izip = zip
quiet = int(quiet)
model = cmd.get_model(selection).atom
x = [i.coord for i in model]
mass = [i.get_mass() for i in model]
xm = [(m*i,m*j,m*k) for (i,j,k),m in izip(x,mass)]
tmass = sum(mass)
rr = sum(mi*i+mj*j+mk*k for (i,j,k),(mi,mj,mk) in izip(x,xm))
mm = sum((sum(i)/tmass)**2 for i in izip(*xm))
rg = math.sqrt(rr/tmass - mm)
if not quiet:
print("Radius of gyration: %.2f" % (rg))
return rg
cmd.extend("rgyrate", rgyrate)
# vi:expandtab
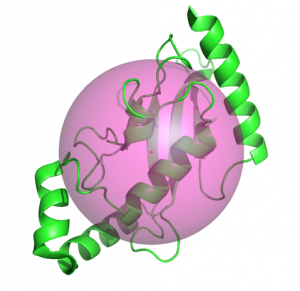
Example
This example uses the radius of gyration and the center of mass to display a semitransparent sphere.
from pymol import cmd, stored, util
import centerOfMass, radiusOfGyration
cmd.set('sphere_transparency', 0.5)
cmd.fetch('2xwu')
cmd.hide('everything')
cmd.show('cartoon', 'chain A')
r = radiusOfGyration.rgyrate('chain A and polymer')
centerOfMass.com('chain A and polymer', object='com', vdw=r)
util.cbc()