ResDe

ResDe is a suite of programs designed to assist crystallographers in creating user defined hydrogen bond distance restraints, which can be helpful when refining low-resolution structures. ResDe is implemented in Python and uses the PyMOL and Tk graphical interfaces.
Overview
The ResDe suite consists of four tools designed to create, edit, and manage user defined hydrogen bond restraints. Restraint Define and Restraint Checker use the PyMOL graphical interface while Restraint Compare and Modify PDB are stand-alone programs using Tk.
Download
NEW: ResDe0.5 Released September 19, 2010! Update your version!
The ResDe suite was developed in the lab of Dr. Sean Johnson and can be downloaded on the ResDe project page.
The ResDe Suite
Restraint Definer
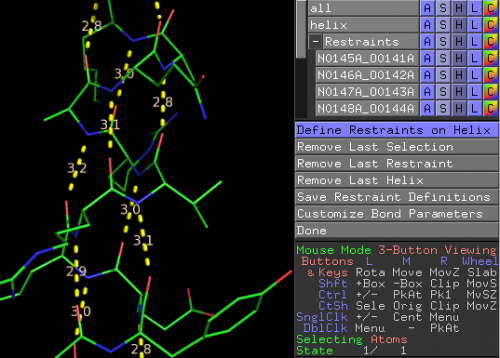
Restraint Definer is a PyMol plugin. After installation the user can access this tool by simply using the plugin menu in PyMol. The user will first open their structure in PyMol. Restraint Definer has three modes; 'Single Restraint', 'Helix (click)', 'Helix (range)'.
TO CHANGE RESTRAINT DEFINITION MODE: ResDe's Restraint Definer menu is in the right hand column of PyMol. The top button in this menu is the 'Mode' button; by default it reads 'Define Single Restraints'. clicking on this button will cause a drop menu to appear from which the user can select either 'Define Single Restraint', 'Define Helix (click)', 'Define Helix (range)' mode.
Upon running Restraint Definer (via the Plugin menu), the program will be in 'Single Restraint' mode by default. To define a restraint in this mode, the user simply clicks on the two atoms they wish to restrain. This will cause a dotted line to form between the two atoms (this is simply a variation of PyMol's measurement tool). When in 'Helix (click)' mode, the user clicks on the two helix termini residues (any atom), order does not matter. Restraints (dotted lines) will appear between the carbonyl oxygens and the n+4 backbone nitrogens along the entire helix. When in 'Helix (range)' mode, a widow comes up asking the user which object to perform the operation on(i.e. if you are working on ABCD.pdb you would choose object ABCD). After selecting the object another widow asks the user about the helix to be defined including chain ID and residue number range. Also, the user can sleek to use a threshold length and enter that length. If a potential restraint in the residue range is longer than the set threshold the restraint will not be created.
Under the 'Undo" menu the user can undo previous actions; remove last restraint and remove last helix. 'Customize Bond Parameters' allows the user to edit the ideal bond length, slack, and sigma from their default values 2.9, 0 , 0.02, respectively. 'Restrain All Helices' brings up a window where the user can dictate how the operation to define all helices is handled. This includes the threshold setting as explained above (see 'Helix (range) mode). Also, the user can select to run DSS, PyMOL's internal secondary structure assignment algorithm.
As restraints are added, a new collapsable object menu will appear in PyMol called 'Restraints'. This menu contains all restraints that have been defined by the user. Inevitably wrong atoms will be clicked leading to inappropriate restraints. These can be removed by deleting the restraint object (the dotted line) under the Restraints menu (the objects are named using the residue numbers and atom names of the two atoms that are restrained).
After defining restraints the user can save the restraints by clicking on the 'Save Restraint Definitions' button. ResDe will ask the user which restraint format that they wish to save their file in, Phenix or ccp4. This restraint definition file is ready to be used in structure refinement using the respective refinement software. If you don't complete defining restraints in one session you can save the restraint definitions and bring them up later (using Restraint Checker) to add more restraints.
Restraint Checker
Restraint Checker is a PyMol plugin. After installation the user can access this tool by simply using the plugin menu in PyMol. The user will first open their structure in PyMol. Upon running Restraint Checker (via the Plugin menu), the user will first be asked what format their restraint file is (phenix or ccp4). The user then opens the restraint file that they want to display. The user is asked to choose the PyMOl object to display the restraints on (this is essential for the program and is useful if there is more than one object open in PyMol). Like Restraint Definer, a dotted line will appear between each pair of atoms that have been restrained. Also like Restraint Definer, the collapsible object menu called 'Restraints' will appear where all the restraint objects are listed.
This tool is useful if the user has a restraint definition file that they would like modify in some way. To make modifications the user will use Restraint Checker to bring up previously defined restraints and then use Restraint Definer to make the desired modifications. For example, the user may have a restraint definition file that they would like to add more restraints to. If the user wants to eliminate a previously defined restraint, they can remove the restraint by identifying and deleting the restraint object in the 'Restraint' object menu. After the modifications have been made the user can save a new restraint definition file.
NOTE: If the user has previously defined restraints on their structure and then runs Restraints Definer, the previously defined restraints will turn red while all newly defined restraints will be yellow.
Restraint Compare
Restrain Compare is a standalone program that is run by simply opening the program. There are special requirements depending on the platform that you are using (see installation instructions). Upon running Restraint Compare the user will be asked to open two different restraint definition files. The user is then asked to confirm that they have opened the correct files and are asked to select which format the files are in (Phenix or ccp4). They are given the option to change the file if needed. The two files are referred to as 'File1' and 'File2'. The user then can view the differences which will come up in a separate window with the option to save the differences as a txt file. The differences that are shown are restraint unique to one file and restraints that restrain identical atoms but have differing restraint parameters.
Modify PDB
Modify PDB is a stand-alone program that is run by simply opening the program or running it from the command line. There are special requirements depending on the platform that you are using (see installation instructions). Upon running Modify PDB the user will be asked to open the PDB file that was used to create the original restraint definition file. The user can then modify the PDB by either offsetting residue numbers or changing chain IDs.
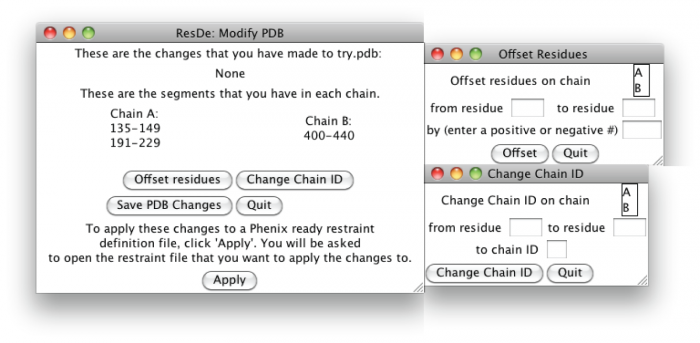
When offsetting residue numbers the user will be asked to provide the chain ID and the region of residues numbers that they wish to modify. They will also be asked to provide the number by which to offset the region by; the number can be either positive or negative.
When changing chain IDs the user will be asked to provide the chain ID and the region of residues numbers that they wish to modify. They will also be asked to provide the new chain ID.
The user can save a new PDB with the changes that they have made by selecting 'Save PDB Changes.'
To apply these changes to an existing restraint definition file the user will click 'Apply' and be asked to open the restraint definition file. The user then is asked to save the new restraint definition file.