CAVER 2.0 is a software tool for protein analysis leading to detection of channels. Channels are in fact void pathways leading from buried cavities to the outside solvent of a protein structure. Studying of these pathways is highly important in the area of drug design and molecular enzymology.
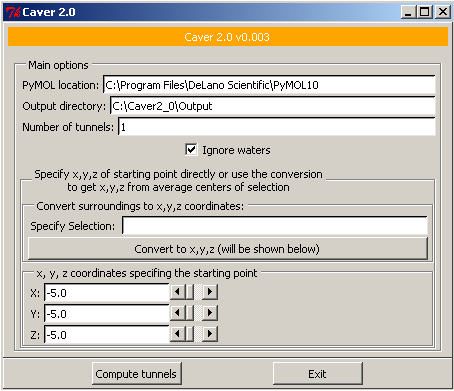
CAVER 2.0 provides rapid, accurate and fully automated calculation of channels not only for static molecules, but also for dynamic molecular simulations.
CAVER 2.0 facilitates analysis of any molecular structure including proteins, nucleic acids or inorganic materials.

CAVER 2.0 can be used in two possible ways. If you are accustomed with the PyMOL ..→


